Lethal variants in humans: lessons learned from a large molecular autopsy cohort
- PMID: 34645488
- PMCID: PMC8511862
- DOI: 10.1186/s13073-021-00973-0
Lethal variants in humans: lessons learned from a large molecular autopsy cohort
Abstract
Background: Molecular autopsy refers to DNA-based identification of the cause of death. Despite recent attempts to broaden its scope, the term remains typically reserved to sudden unexplained death in young adults. In this study, we aim to showcase the utility of molecular autopsy in defining lethal variants in humans.
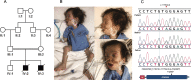
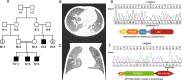
Methods: We describe our experience with a cohort of 481 cases in whom the cause of premature death was investigated using DNA from the index or relatives (molecular autopsy by proxy). Molecular autopsy tool was typically exome sequencing although some were investigated using targeted approaches in the earlier stages of the study; these include positional mapping, targeted gene sequencing, chromosomal microarray, and gene panels.
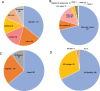
Results: The study includes 449 cases from consanguineous families and 141 lacked family history (simplex). The age range was embryos to 18 years. A likely causal variant (pathogenic/likely pathogenic) was identified in 63.8% (307/481), a much higher yield compared to the general diagnostic yield (43%) from the same population. The predominance of recessive lethal alleles allowed us to implement molecular autopsy by proxy in 55 couples, and the yield was similarly high (63.6%). We also note the occurrence of biallelic lethal forms of typically non-lethal dominant disorders, sometimes representing a novel bona fide biallelic recessive disease trait. Forty-six disease genes with no OMIM phenotype were identified in the course of this study. The presented data support the candidacy of two other previously reported novel disease genes (FAAH2 and MSN). The focus on lethal phenotypes revealed many examples of interesting phenotypic expansion as well as remarkable variability in clinical presentation. Furthermore, important insights into population genetics and variant interpretation are highlighted based on the results.
Conclusions: Molecular autopsy, broadly defined, proved to be a helpful clinical approach that provides unique insights into lethal variants and the clinical annotation of the human genome.
Keywords: BMPR1A; EHBP1L1; Embryonic lethal; FAAH2; MSN; founder; loss of function; multi-locus.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Ackerman, M.J., Tester, D.J., and Porter, C.B.J. (1999). Swimming, a gene-specific arrhythmogenic trigger for inherited long QT syndrome. In Mayo Clinic Proceedings. (Elsevier), pp 1088-1094. - PubMed
-
- Quinlan-Jones E, Lord J, Williams D, Hamilton S, Marton T, Eberhardt RY, et al. Molecular autopsy by trio exome sequencing (ES) and postmortem examination in fetuses and neonates with prenatally identified structural anomalies. Genet Med. 2019;21:1065–1073. doi: 10.1038/s41436-018-0298-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

