Microbial colonization and resistome dynamics in food processing environments of a newly opened pork cutting industry during 1.5 years of activity
- PMID: 34645520
- PMCID: PMC8515711
- DOI: 10.1186/s40168-021-01131-9
Microbial colonization and resistome dynamics in food processing environments of a newly opened pork cutting industry during 1.5 years of activity
Abstract
Background: The microorganisms that inhabit food processing environments (FPE) can strongly influence the associated food quality and safety. In particular, the possibility that FPE may act as a reservoir of antibiotic-resistant microorganisms, and a hotspot for the transmission of antibiotic resistance genes (ARGs) is a concern in meat processing plants. Here, we monitor microbial succession and resistome dynamics relating to FPE through a detailed analysis of a newly opened pork cutting plant over 1.5 years of activity.
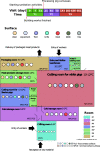
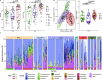
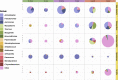
Results: We identified a relatively restricted principal microbiota dominated by Pseudomonas during the first 2 months, while a higher taxonomic diversity, an increased representation of other taxa (e.g., Acinetobacter, Psychrobacter), and a certain degree of microbiome specialization on different surfaces was recorded later on. An increase in total abundance, alpha diversity, and β-dispersion of ARGs, which were predominantly assigned to Acinetobacter and associated with resistance to certain antimicrobials frequently used on pig farms of the region, was detected over time. Moreover, a sharp increase in the occurrence of extended-spectrum β-lactamase-producing Enterobacteriaceae and vancomycin-resistant Enterococcaceae was observed when cutting activities started. ARGs associated with resistance to β-lactams, tetracyclines, aminoglycosides, and sulphonamides frequently co-occurred, and mobile genetic elements (i.e., plasmids, integrons) and lateral gene transfer events were mainly detected at the later sampling times in drains.
Conclusions: The observations made suggest that pig carcasses were a source of resistant bacteria that then colonized FPE and that drains, together with some food-contact surfaces, such as equipment and table surfaces, represented a reservoir for the spread of ARGs in the meat processing facility. Video Abstract.
Keywords: Antimicrobial resistance; Food processing environments; Metagenomics; Microbial ecology.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Larsen MH, Dalmasso M, Ingmer H, Langsrud S, Malakauskas M, Mader A, Møretrø T, Smole Možina S, Rychli K, Wagner M, John Wallace R, Zentek J, Jordan K. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control. 2014;44:92–109. doi: 10.1016/j.foodcont.2014.03.039. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

