Differential impact of a dyskeratosis congenita mutation in TPP1 on mouse hematopoiesis and germline
- PMID: 34645668
- PMCID: PMC8548261
- DOI: 10.26508/lsa.202101208
Differential impact of a dyskeratosis congenita mutation in TPP1 on mouse hematopoiesis and germline
Abstract
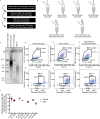
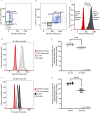
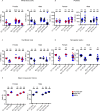
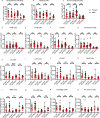
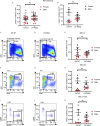
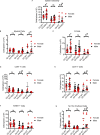
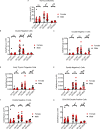
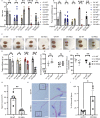
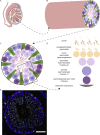
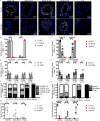
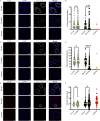
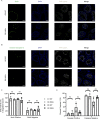
Telomerase extends chromosome ends in somatic and germline stem cells to ensure continued proliferation. Mutations in genes critical for telomerase function result in telomeropathies such as dyskeratosis congenita, frequently resulting in spontaneous bone marrow failure. A dyskeratosis congenita mutation in TPP1 (K170∆) that specifically compromises telomerase recruitment to telomeres is a valuable tool to evaluate telomerase-dependent telomere length maintenance in mice. We used CRISPR-Cas9 to generate a mouse knocked in for the equivalent of the TPP1 K170∆ mutation (TPP1 K82∆) and investigated both its hematopoietic and germline compartments in unprecedented detail. TPP1 K82∆ caused progressive telomere erosion with increasing generation number but did not induce steady-state hematopoietic defects. Strikingly, K82∆ caused mouse infertility, consistent with gross morphological defects in the testis and sperm, the appearance of dysfunctional seminiferous tubules, and a decrease in germ cells. Intriguingly, both TPP1 K82∆ mice and previously characterized telomerase knockout mice show no spontaneous bone marrow failure but rather succumb to infertility at steady-state. We speculate that telomere length maintenance contributes differently to the evolutionary fitness of humans and mice.
© 2021 Graniel et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- T32 AG000114/AG/NIA NIH HHS/United States
- T32 HL007439/HL/NHLBI NIH HHS/United States
- F30 HD097961/HD/NICHD NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- DP2 HD091949/HD/NICHD NIH HHS/United States
- T32 AI070077/AI/NIAID NIH HHS/United States
- T32 CA009140/CA/NCI NIH HHS/United States
- T32 HD079342/HD/NICHD NIH HHS/United States
- T32 GM007863/GM/NIGMS NIH HHS/United States
- R25 GM086262/GM/NIGMS NIH HHS/United States
- R01 HD092084/HD/NICHD NIH HHS/United States
- R01 AG050509/AG/NIA NIH HHS/United States
- R01 GM120094/GM/NIGMS NIH HHS/United States
- F30 AI136315/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
