Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance
- PMID: 34645820
- PMCID: PMC8514477
- DOI: 10.1038/s41467-021-26266-z
Specific gut microbiome signatures and the associated pro-inflamatory functions are linked to pediatric allergy and acquisition of immune tolerance
Abstract
Understanding the functional potential of the gut microbiome is of primary importance for the design of innovative strategies for allergy treatment and prevention. Here we report the gut microbiome features of 90 children affected by food (FA) or respiratory (RA) allergies and 30 age-matched, healthy controls (CT). We identify specific microbial signatures in the gut microbiome of allergic children, such as higher abundance of Ruminococcus gnavus and Faecalibacterium prausnitzii, and a depletion of Bifidobacterium longum, Bacteroides dorei, B. vulgatus and fiber-degrading taxa. The metagenome of allergic children shows a pro-inflammatory potential, with an enrichment of genes involved in the production of bacterial lipo-polysaccharides and urease. We demonstrate that specific gut microbiome signatures at baseline can be predictable of immune tolerance acquisition. Finally, a strain-level selection occurring in the gut microbiome of allergic subjects is identified. R. gnavus strains enriched in FA and RA showed lower ability to degrade fiber, and genes involved in the production of a pro-inflammatory polysaccharide. We demonstrate that a gut microbiome dysbiosis occurs in allergic children, with R. gnavus emerging as a main player in pediatric allergy. These findings may open new strategies in the development of innovative preventive and therapeutic approaches. Trial: NCT04750980.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
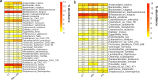

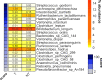



References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources
Medical

