The network interplay of interferon and Toll-like receptor signaling pathways in the anti-Candida immune response
- PMID: 34645905
- PMCID: PMC8514550
- DOI: 10.1038/s41598-021-99838-0
The network interplay of interferon and Toll-like receptor signaling pathways in the anti-Candida immune response
Abstract
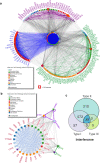
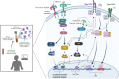
Fungal infections represent a major global health problem affecting over a billion people that kills more than 1.5 million annually. In this study, we employed an integrative approach to reveal the landscape of the human immune responses to Candida spp. through meta-analysis of microarray, bulk, and single-cell RNA sequencing (scRNA-seq) data for the blood transcriptome. We identified across these different studies a consistent interconnected network interplay of signaling molecules involved in both Toll-like receptor (TLR) and interferon (IFN) signaling cascades that is activated in response to different Candida species (C. albicans, C. auris, C. glabrata, C. parapsilosis, and C. tropicalis). Among these molecules are several types I IFN, indicating an overlap with antiviral immune responses. scRNA-seq data confirmed that genes commonly identified by the three transcriptomic methods show cell type-specific expression patterns in various innate and adaptive immune cells. These findings shed new light on the anti-Candida immune response, providing putative molecular pathways for therapeutic intervention.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Signaling through Syk or CARD9 Mediates Species-Specific Anti-Candida Protection in Bone Marrow Chimeric Mice.mBio. 2021 Aug 31;12(4):e0160821. doi: 10.1128/mBio.01608-21. Epub 2021 Aug 31. mBio. 2021. PMID: 34465030 Free PMC article.
-
Conventional dendritic cells mount a type I IFN response against Candida spp. requiring novel phagosomal TLR7-mediated IFN-β signaling.J Immunol. 2011 Mar 1;186(5):3104-12. doi: 10.4049/jimmunol.1002599. Epub 2011 Jan 31. J Immunol. 2011. PMID: 21282509
-
Vaccination with Secreted Aspartyl Proteinase 2 Protein from Candida parapsilosis Can Enhance Survival of Mice during C. tropicalis-Mediated Systemic Candidiasis.Infect Immun. 2020 Sep 18;88(10):e00312-20. doi: 10.1128/IAI.00312-20. Print 2020 Sep 18. Infect Immun. 2020. PMID: 32661125 Free PMC article.
-
Innate immune cell response upon Candida albicans infection.Virulence. 2016 Jul 3;7(5):512-26. doi: 10.1080/21505594.2016.1138201. Epub 2016 Apr 14. Virulence. 2016. PMID: 27078171 Free PMC article. Review.
-
Interplay between Candida albicans and the mammalian innate host defense.Infect Immun. 2012 Apr;80(4):1304-13. doi: 10.1128/IAI.06146-11. Epub 2012 Jan 17. Infect Immun. 2012. PMID: 22252867 Free PMC article. Review.
Cited by
-
Candida albicans-enteric viral interactions-The prostaglandin E2 connection and host immune responses.iScience. 2022 Dec 24;26(1):105870. doi: 10.1016/j.isci.2022.105870. eCollection 2023 Jan 20. iScience. 2022. PMID: 36647379 Free PMC article. Review.
-
The emerging role of oral microbiota in oral cancer initiation, progression and stemness.Front Immunol. 2023 Oct 26;14:1198269. doi: 10.3389/fimmu.2023.1198269. eCollection 2023. Front Immunol. 2023. PMID: 37954619 Free PMC article. Review.
-
Immunological signatures unveiled by integrative systems vaccinology characterization of dengue vaccination trials and natural infection.Front Immunol. 2024 Feb 20;15:1282754. doi: 10.3389/fimmu.2024.1282754. eCollection 2024. Front Immunol. 2024. PMID: 38444851 Free PMC article.
-
Occurrence of Candidemia in Patients with COVID-19 Admitted to Five ICUs in France.J Fungi (Basel). 2022 Jun 28;8(7):678. doi: 10.3390/jof8070678. J Fungi (Basel). 2022. PMID: 35887435 Free PMC article.
-
Nakaseomyces glabratus drug resistance genes expression in vulvovaginal candidiasis: a systematic review.Curr Med Mycol. 2024 Feb 1;10:e2024.345248.1573. doi: 10.22034/cmm.2024.345248.1573. eCollection 2024. Curr Med Mycol. 2024. PMID: 40584607 Free PMC article. Review.
References
-
- World Health Organization. GLASS report: early implementation 2017–2018. https://www.who.int/publications/i/item/9789241515061 (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

