Mutant clones in normal epithelium outcompete and eliminate emerging tumours
- PMID: 34646013
- PMCID: PMC7612642
- DOI: 10.1038/s41586-021-03965-7
Mutant clones in normal epithelium outcompete and eliminate emerging tumours
Abstract
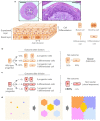
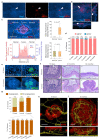
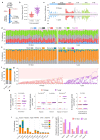
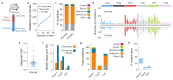
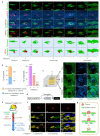
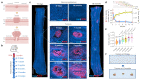
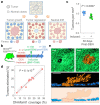
Human epithelial tissues accumulate cancer-driver mutations with age1-9, yet tumour formation remains rare. The positive selection of these mutations suggests that they alter the behaviour and fitness of proliferating cells10-12. Thus, normal adult tissues become a patchwork of mutant clones competing for space and survival, with the fittest clones expanding by eliminating their less competitive neighbours11-14. However, little is known about how such dynamic competition in normal epithelia influences early tumorigenesis. Here we show that the majority of newly formed oesophageal tumours are eliminated through competition with mutant clones in the adjacent normal epithelium. We followed the fate of nascent, microscopic, pre-malignant tumours in a mouse model of oesophageal carcinogenesis and found that most were rapidly lost with no indication of tumour cell death, decreased proliferation or an anti-tumour immune response. However, deep sequencing of ten-day-old and one-year-old tumours showed evidence of selection on the surviving neoplasms. Induction of highly competitive clones in transgenic mice increased early tumour removal, whereas pharmacological inhibition of clonal competition reduced tumour loss. These results support a model in which survival of early neoplasms depends on their competitive fitness relative to that of mutant clones in the surrounding normal tissue. Mutant clones in normal epithelium have an unexpected anti-tumorigenic role in purging early tumours through cell competition, thereby preserving tissue integrity.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures






Comment in
-
Clone competition as a mechanism to reduce tumor formation.Dev Cell. 2021 Dec 20;56(24):3307-3308. doi: 10.1016/j.devcel.2021.12.003. Dev Cell. 2021. PMID: 34932947
-
Cell competition: Clonal competition protects against early tumorigenesis.Curr Biol. 2022 Jan 10;32(1):R52-R54. doi: 10.1016/j.cub.2021.11.029. Curr Biol. 2022. PMID: 35015999
References
-
- Lee-Six H, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. - PubMed
-
- Suda K, et al. Clonal Expansion and Diversification of Cancer-Associated Mutations in Endometriosis and Normal Endometrium. Cell reports. 2018;24:1777–1789. - PubMed
-
- Yokoyama A, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

