Analysis of the Expression and Prognostic Potential of a Novel Metabolic Regulator ANGPTL8/Betatrophin in Human Cancers
- PMID: 34646087
- PMCID: PMC8502826
- DOI: 10.3389/pore.2021.1609914
Analysis of the Expression and Prognostic Potential of a Novel Metabolic Regulator ANGPTL8/Betatrophin in Human Cancers
Abstract
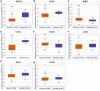
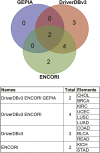
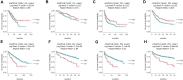
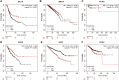
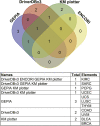
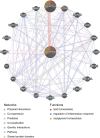
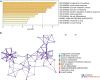
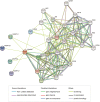
The angiopoietin-like protein (ANGPTL) family members, except for the novel atypical member ANGPTL8/betatrophin, have been reported to participate in angiogenesis, inflammation and cancer. ANGPTL8/betatrophin is a metabolic regulator that is involved in lipid metabolism and glucose homeostasis. However, little is known about the expression and prognostic value of ANGPTL8/betatrophin in human cancers. In this study, we first conducted detailed analyses of ANGPTL8/betatrophin expression in cancer/normal samples via the Human Protein Atlas (HPA), Gene Expression Profiling Interactive Analysis (GEPIA), DriverDBv3, ENCORI and UALCAN databases. ANGPTL8/betatrophin showed high tissue specificity (enriched in the liver) and cell-type specificity (enriched in HepG2 and MCF7 cell lines). More than one databases demonstrated that the gene expression of ANGPTL8/betatrophin was significantly lower in cholangiocarcinoma (CHOL), breast invasive carcinoma (BRCA), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), uterine corpus endometrial carcinoma (UCEC), and significantly higher in kidney renal clear cell carcinoma (KIRC) compared with that in normal samples. However, the protein expression of ANGPTL8/betatrophin displayed opposite results in clear cell renal cell carcinoma (ccRCC)/KIRC. Based on the expression profiles, the prognostic value was evaluated with the GEPIA, DriverDBv3, Kaplan Meier plotter and ENCORI databases. Two or more databases demonstrated that ANGPTL8/betatrophin significantly affected the survival of KIRC, uterine corpus endometrial carcinoma (UCEC), pheochromocytoma and paraganglioma (PCPG) and sarcoma (SARC); patients with PCPG and SARC may benifit from high ANGPTL8/betatrophin expression while high ANGPTL8/betatrophin expression was associated with poor prognosis in KIRC and UCEC. Functional analyses with the GeneMANIA, Metascape and STRING databases suggested that ANGPTL8/betatrophin was mainly involved in lipid homeostasis, especially triglyceride and cholesterol metabolism; glucose homeostasis, especially insulin resistance; AMPK signaling pathway; PI3K/Akt signaling pathway; PPAR signaling pathway; mTOR signaling pathway; HIF-1 signaling pathway; autophagy; regulation of inflammatory response. ANGPTL8/betatrophin may be a promising prognostic biomarker and therapeutic target, thus providing evidence to support further exploration of its role in defined human cancers.
Keywords: ANGPTL8/betatrophin; Akt signaling; differential expression analysis; glucose homeostasis; inflammation; lipid metabolism; prognostic biomarker.
Copyright © 2021 Xu, Tian, Shi, Sun and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
ANGPTL8/betatrophin alleviates insulin resistance via the Akt-GSK3β or Akt-FoxO1 pathway in HepG2 cells.Exp Cell Res. 2016 Jul 15;345(2):158-67. doi: 10.1016/j.yexcr.2015.09.012. Epub 2015 Sep 24. Exp Cell Res. 2016. PMID: 26387753
-
An Integrated Computational Analysis of High-Risk SNPs in Angiopoietin-like Proteins (ANGPTL3 and ANGPTL8) Reveals Perturbed Protein Dynamics Associated with Cancer.Molecules. 2023 Jun 8;28(12):4648. doi: 10.3390/molecules28124648. Molecules. 2023. PMID: 37375208 Free PMC article.
-
Angiopoietin-like protein 8 (betatrophin) inhibits hepatic gluconeogenesis through PI3K/Akt signaling pathway in diabetic mice.Metabolism. 2022 Jan;126:154921. doi: 10.1016/j.metabol.2021.154921. Epub 2021 Oct 27. Metabolism. 2022. PMID: 34715116
-
ANGPTL8 in metabolic homeostasis: more friend than foe?Open Biol. 2021 Sep;11(9):210106. doi: 10.1098/rsob.210106. Epub 2021 Sep 29. Open Biol. 2021. PMID: 34582711 Free PMC article. Review.
-
ANGPTL8 (betatrophin) role in diabetes and metabolic diseases.Diabetes Metab Res Rev. 2017 Nov;33(8). doi: 10.1002/dmrr.2919. Epub 2017 Aug 30. Diabetes Metab Res Rev. 2017. PMID: 28722798 Review.
Cited by
-
ANGPTL8 links inflammation and poor differentiation, which are characteristics of malignant renal cell carcinoma.Cancer Sci. 2023 Apr;114(4):1410-1422. doi: 10.1111/cas.15700. Epub 2023 Jan 2. Cancer Sci. 2023. PMID: 36529524 Free PMC article.
-
Proteomics analysis of carotid body tumor revealed potential mechanisms and molecular differences among Shamblin classifications.Exp Biol Med (Maywood). 2023 Oct;248(20):1785-1798. doi: 10.1177/15353702231199475. Epub 2023 Oct 16. Exp Biol Med (Maywood). 2023. PMID: 37845830 Free PMC article.
-
Dual role of ANGPTL8 in promoting tumor cell proliferation and immune escape during hepatocarcinogenesis.Oncogenesis. 2023 May 15;12(1):26. doi: 10.1038/s41389-023-00473-3. Oncogenesis. 2023. PMID: 37188659 Free PMC article.
-
Emerging insights into the roles of ANGPTL8 beyond glucose and lipid metabolism.Front Physiol. 2023 Dec 1;14:1275485. doi: 10.3389/fphys.2023.1275485. eCollection 2023. Front Physiol. 2023. PMID: 38107478 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

