Identification of Novel Diagnosis Biomarkers for Therapy-Related Neuroendocrine Prostate Cancer
- PMID: 34646089
- PMCID: PMC8503838
- DOI: 10.3389/pore.2021.1609968
Identification of Novel Diagnosis Biomarkers for Therapy-Related Neuroendocrine Prostate Cancer
Abstract
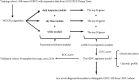
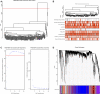
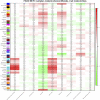
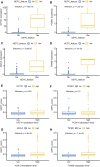
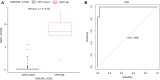
Background: Therapy-related neuroendocrine prostate cancer (NEPC) is a lethal castration-resistant prostate cancer (CRPC) subtype that, at present, lacks well-characterized molecular biomarkers. The clinical diagnosis of this disease is dependent on biopsy and histological assessment: methods that are experience-based and easily misdiagnosed due to tumor heterogeneity. The development of robust diagnostic tools for NEPC may assist clinicians in making medical decisions on the choice of continuing anti-androgen receptor therapy or switching to platinum-based chemotherapy. Methods: Gene expression profiles and clinical characteristics data of 208 samples of metastatic CRPC, including castration-resistant prostate adenocarcinoma (CRPC-adeno) and castration-resistant neuroendocrine prostate adenocarcinoma (CRPC-NE), were obtained from the prad_su2c_2019 dataset. Weighted Gene Co-expression Network Analysis (WGCNA) was subsequently used to construct a free-scale gene co-expression network to study the interrelationship between the potential modules and clinical features of metastatic prostate adenocarcinoma and to identify hub genes in the modules. Furthermore, the least absolute shrinkage and selection operator (LASSO) regression analysis was used to build a model to predict the clinical characteristics of CRPC-NE. The findings were then verified in the nepc_wcm_2016 dataset. Results: A total of 51 co-expression modules were successfully constructed using WGCNA, of which three co-expression modules were found to be significantly associated with the neuroendocrine features and the NEPC score. In total, four novel genes, including NPTX1, PCSK1, ASXL3, and TRIM9, were all significantly upregulated in NEPC compared with the adenocarcinoma samples, and these genes were all associated with the neuroactive ligand receptor interaction pathway. Next, the expression levels of these four genes were used to construct an NEPC diagnosis model, which was successfully able to distinguish CRPC-NE from CRPC-adeno samples in both the training and the validation cohorts. Moreover, the values of the area under the receiver operating characteristic (AUC) were 0.995 and 0.833 for the training and validation cohorts, respectively. Conclusion: The present study identified four specific novel biomarkers for therapy-related NEPC, and these biomarkers may serve as an effective tool for the diagnosis of NEPC, thereby meriting further study.
Keywords: LASSO; WGCNA; biomarker; mCRPC; neuroendocrine prostate cancer (NEPC); signature.
Copyright © 2021 Zhang, Qian, Wu, Zhu, Yu, Gong, Li, He and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- MaryBeth BC, Isabelle S, Jason AE, Freddie B, Ahmedin J. Recent Global Patterns in Prostate Cancer Incidence and Mortality Rates. Amsterdam, Netharlands: European Urology; (2019). - PubMed
-
- Vincenza C, Clara O, Kenneth WE, Rohan B, Michael S, Ana M, et al. Clinical Features of Neuroendocrine Prostate Cancer. Eur J Cancer (2019).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

