Adintoviruses: a proposed animal-tropic family of midsize eukaryotic linear dsDNA (MELD) viruses
- PMID: 34646575
- PMCID: PMC8502044
- DOI: 10.1093/ve/veaa055
Adintoviruses: a proposed animal-tropic family of midsize eukaryotic linear dsDNA (MELD) viruses
Abstract
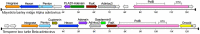
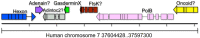
Polintons (also known as Mavericks) were initially identified as a widespread class of eukaryotic transposons named for their hallmark type B DNA polymerase and retrovirus-like integrase genes. It has since been recognized that many polintons encode possible capsid proteins and viral genome-packaging ATPases similar to those of a diverse range of double-stranded DNA viruses. This supports the inference that at least some polintons are actually viruses capable of cell-to-cell spread. At present, there are no polinton-associated capsid protein genes annotated in public sequence databases. To rectify this deficiency, we used a data-mining approach to investigate the distribution and gene content of polinton-like elements and related DNA viruses in animal genomic and metagenomic sequence datasets. The results define a discrete family-like clade of viruses with two genus-level divisions. We propose the family name Adintoviridae, connoting similarities to adenovirus virion proteins and the presence of a retrovirus-like integrase gene. Although adintovirus-class PolB sequences were detected in datasets for fungi and various unicellular eukaryotes, sequences resembling adintovirus virion proteins and accessory genes appear to be restricted to animals. Degraded adintovirus sequences are endogenized into the germlines of a wide range of animals, including humans.
Keywords: Maverick; adenain; capsid; polB; polinton; recombination; transposon.
Published by Oxford University Press 2020. This work is written by a US Government employee and is in the public domain in the US.
Figures





References
LinkOut - more resources
Full Text Sources

