circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing
- PMID: 34647522
- PMCID: PMC8550772
- DOI: 10.7554/eLife.69457
circFL-seq reveals full-length circular RNAs with rolling circular reverse transcription and nanopore sequencing
Abstract
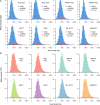
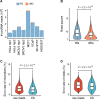
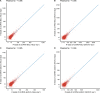
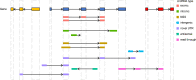
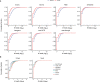
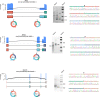
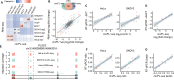
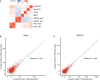
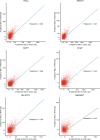
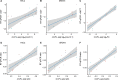
Circular RNAs (circRNAs) act through multiple mechanisms via their sequence features to fine-tune gene expression networks. Due to overlapping sequences with linear cognates, identifying internal sequences of circRNAs remains a challenge, which hinders a comprehensive understanding of circRNA functions and mechanisms. Here, based on rolling circular reverse transcription and nanopore sequencing, we developed circFL-seq, a full-length circRNA sequencing method, to profile circRNA at the isoform level. With a customized computational pipeline to directly identify full-length sequences from rolling circular reads, we reconstructed 77,606 high-quality circRNAs from seven human cell lines and two human tissues. circFL-seq benefits from rolling circles and long-read sequencing, and the results showed more than tenfold enrichment of circRNA reads and advantages for both detection and quantification at the isoform level compared to those for short-read RNA sequencing. The concordance of the RT-qPCR and circFL-seq results for the identification of differential alternative splicing suggested wide application prospects for functional studies of internal variants in circRNAs. Moreover, the detection of fusion circRNAs at the omics scale may further expand the application of circFL-seq. Taken together, the accurate identification and quantification of full-length circRNAs make circFL-seq a potential tool for large-scale screening of functional circRNAs.
Keywords: chromosomes; circular RNA; computational biology; full-length sequencing; fusion circRNA; gene expression; human; nanopore sequencing; systems biology.
© 2021, Liu et al.
Conflict of interest statement
ZL, CT, SL, MD, YB, XH, YL, JC, EY No competing interests declared
Figures





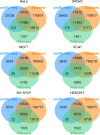
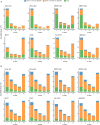
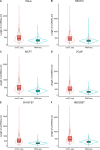
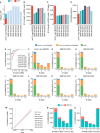




References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

