Obesity Impairs Embryonic Myogenesis by Enhancing BMP Signaling within the Dermomyotome
- PMID: 34647690
- PMCID: PMC8596142
- DOI: 10.1002/advs.202102157
Obesity Impairs Embryonic Myogenesis by Enhancing BMP Signaling within the Dermomyotome
Abstract
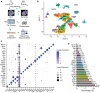
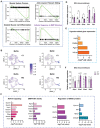
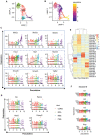
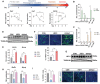
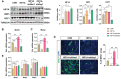
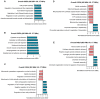
Obesity during pregnancy leads to adverse health outcomes in offspring. However, the initial effects of maternal obesity (MO) on embryonic organogenesis have yet to be thoroughly examined. Using unbiased single-cell transcriptomic analyses (scRNA-seq), the effects of MO on the myogenic process is investigated in embryonic day 9.5 (E9.5) mouse embryos. The results suggest that MO induces systematic hypoxia, which is correlated with enhanced BMP signaling and impairs skeletal muscle differentiation within the dermomyotome (DM). The Notch-signaling effectors, HES1 and HEY1, which also act down-stream of BMP signaling, suppress myogenic differentiation through transcriptionally repressing the important myogenic regulator MEF2C. Moreover, the major hypoxia effector, HIF1A, enhances expression of HES1 and HEY1 and blocks myogenic differentiation in vitro. In summary, this data demonstrate that MO induces hypoxia and impairs myogenic differentiation by up-regulating BMP signaling within the DM, which may account for the disruptions of skeletal muscle development and function in progeny.
Keywords: bone morphogenetic proteins signaling; embryonic myogenesis; maternal obesity; single cell RNA sequencing.
© 2021 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Poston L., Caleyachetty R., Cnattingius S., Corvalán C., Uauy R., Herring S., Gillman M. W., Lancet Diabetes Endocrinol. 2016, 4, 1025. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
