Cardiac SARS-CoV-2 infection is associated with pro-inflammatory transcriptomic alterations within the heart
- PMID: 34647998
- PMCID: PMC8803085
- DOI: 10.1093/cvr/cvab322
Cardiac SARS-CoV-2 infection is associated with pro-inflammatory transcriptomic alterations within the heart
Abstract
Aims: Cardiac involvement in COVID-19 is associated with adverse outcome. However, it is unclear whether cell-specific consequences are associated with cardiac SARS-CoV-2 infection. Therefore, we investigated heart tissue utilizing in situ hybridization, immunohistochemistry, and RNA-sequencing in consecutive autopsy cases to quantify virus load and characterize cardiac involvement in COVID-19.
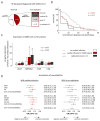
Methods and results: In this study, 95 SARS-CoV-2-positive autopsy cases were included. A relevant SARS-CoV-2 virus load in the cardiac tissue was detected in 41/95 deceased (43%). Massive analysis of cDNA ends (MACE)-RNA-sequencing was performed to identify molecular pathomechanisms caused by the infection of the heart. A signature matrix was generated based on the single-cell dataset 'Heart Cell Atlas' and used for digital cytometry on the MACE-RNA-sequencing data. Thus, immune cell fractions were estimated and revealed no difference in immune cell numbers in cases with and without cardiac infection. This result was confirmed by quantitative immunohistological diagnosis. MACE-RNA-sequencing revealed 19 differentially expressed genes (DEGs) with a q-value <0.05 (e.g. up: IFI44L, IFT3, TRIM25; down: NPPB, MB, MYPN). The upregulated DEGs were linked to interferon pathways and originate predominantly from endothelial cells. In contrast, the downregulated DEGs originate predominately from cardiomyocytes. Immunofluorescent staining showed viral protein in cells positive for the endothelial marker ICAM1 but rarely in cardiomyocytes. The Gene Ontology (GO) term analysis revealed that downregulated GO terms were linked to cardiomyocyte structure, whereas upregulated GO terms were linked to anti-virus immune response.
Conclusion: This study reveals that cardiac infection induced transcriptomic alterations mainly linked to immune response and destruction of cardiomyocytes. While endothelial cells are primarily targeted by the virus, we suggest cardiomyocyte destruction by paracrine effects. Increased pro-inflammatory gene expression was detected in SARS-CoV-2-infected cardiac tissue but no increased SARS-CoV-2 associated immune cell infiltration was observed.
Keywords: COVID-19; Cardiac infection; Cardiac signature matrix; MACE; RNA-seq; SARS-CoV-2.
© The Author(s) 2021. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures






References
-
- RECOVERY Collaborative Group, Horby P, Lim WS, Emberson JR, Mafham M, Bell JL, Linsell L, Staplin N, Brightling C, Ustianowski A, Elmahi E, Prudon B, Green C, Felton T, Chadwick D, Rege K, Fegan C, Chappell LC, Faust SN, Jaki T, Jeffery K, Montgomery A, Rowan K, Juszczak E, Baillie JK, Haynes R, Landray MJ. Dexamethasone in hospitalized patients with COVID-19. N Engl J Med 2021;384:693–704. - PMC - PubMed
-
- Puelles VG, Lutgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schroder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 2020;383:590–592. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

