Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India
- PMID: 34648303
- PMCID: PMC7612010
- DOI: 10.1126/science.abj9932
Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India
Abstract
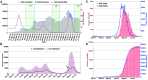
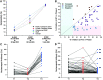
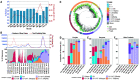
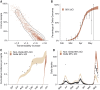
Delhi, the national capital of India, experienced multiple severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) outbreaks in 2020 and reached population seropositivity of >50% by 2021. During April 2021, the city became overwhelmed by COVID-19 cases and fatalities, as a new variant, B.1.617.2 (Delta), replaced B.1.1.7 (Alpha). A Bayesian model explains the growth advantage of Delta through a combination of increased transmissibility and reduced sensitivity to immune responses generated against earlier variants (median estimates: 1.5-fold greater transmissibility and 20% reduction in sensitivity). Seropositivity of an employee and family cohort increased from 42% to 87.5% between March and July 2021, with 27% reinfections, as judged by increased antibody concentration after a previous decline. The likely high transmissibility and partial evasion of immunity by the Delta variant contributed to an overwhelming surge in Delhi.
Figures
References
-
- Murhekar M. V., Bhatnagar T., Thangaraj J. W. V., Saravanakumar V., Kumar M. S., Selvaraju S., Rade K., Kumar C. P. G., Sabarinathan R., Turuk A., Asthana S., Balachandar R., Bangar S. D., Bansal A. K., Chopra V., Das D., Deb A. K., Devi K. R., Dhikav V., Dwivedi G. R., Khan S. M. S., Kumar M. S., Laxmaiah A., Madhukar M., Mahapatra A., Rangaraju C., Turuk J., Yadav R., Andhalkar R., Arunraj K., Bharadwaj D. K., Bharti P., Bhattacharya D., Bhat J., Chahal A. S., Chakraborty D., Chaudhury A., Deval H., Dhatrak S., Dayal R., Elantamilan D., Giridharan P., Haq I., Hudda R. K., Jagjeevan B., Kalliath A., Kanungo S., Krishnan N. N., Kshatri J. S., Kumar A., Kumar N., Kumar V. G. V., Lakshmi G. G. J. N., Mehta G., Mishra N. K., Mitra A., Nagbhushanam K., Nimmathota A., Nirmala A. R., Pandey A. K., Prasad G. V., Qurieshi M. A., Reddy S. D., Robinson A., Sahay S., Saxena R., Sekar K., Shukla V. K., Singh H. B., Singh P. K., Singh P., Singh R., Srinivasan N., Varma D. S., Viramgami A., Wilson V. C., Yadav S., Yadav S., Zaman K., Chakrabarti A., Das A., Dhaliwal R. S., Dutta S., Kant R., Khan A. M., Narain K., Narasimhaiah S., Padmapriyadarshini C., Pandey K., Pati S., Patil S., Rajkumar H., Ramarao T., Sharma Y. K., Singh S., Panda S., Reddy D. C. S., Bhargava B., Anand T., Babu G. R., Chauhan H., Dikid T., Gangakhedkar R. R., Kant S., Kulkarni S., Muliyil J. P., Pandey R. M., Sarkar S., Shah N., Shrivastava A., Singh S. K., Zodpe S., Hindupur A., Asish P. R., Chellakumar M., Chokkalingam D., Dasgupta S., Gowtham M. M. E., Jose A., Kalaiyarasi K., Karthik N. N., Karunakaran T., Kiruthika G., Dinesh Kumar H., Sarath Kumar S., Sarath Kumar M. P., Michaelraj E., Pradhan J., Arun Prasath E. B., Gladys Angelin Rachel D., Rani S., Rozario A., Sivakumar R., Gnana Soundari P., Sujeetha K., Vinod A.; ICMR Serosurveillance Group , SARS-CoV-2 seroprevalence among the general population and healthcare workers in India, December 2020-January 2021. Int. J. Infect. Dis. 108, 145–155 (2021). 10.1016/j.ijid.2021.05.040 - DOI - PMC - PubMed
-
- S. Jain (@SatyendarJain), Twitter post, 2 February 2021, 5:19 a.m.; https://twitter.com/satyendarjain/status/1356547817427226624?lang=en.
-
- Government of National Capital Territory of Delhi, Orders F.23/Misc/COVID-19/DGHS/NHC/2020/Pt XVIII/90-97 and No.01/H&FW/COVID-Cell/2020/07/ss4hfw/726, 12 April 2021; http://health.delhigovt.nic.in/wps/wcm/connect/doit_health/Health/Home/C....
-
- Naushin S., Sardana V., Ujjainiya R., Bhatheja N., Kutum R., Bhaskar A. K., Pradhan S., Prakash S., Khan R., Rawat B. S., Tallapaka K. B., Anumalla M., Chandak G. R., Lahiri A., Kar S., Mulay S. R., Mugale M. N., Srivastava M., Khan S., Srivastava A., Tomar B., Veerapandian M., Venkatachalam G., Vijayakumar S. R., Agarwal A., Gupta D., Halami P. M., Peddha M. S., Sundaram G. M., Veeranna R. P., Pal A., Agarwal V. K., Maurya A. K., Singh R. K., Raman A. K., Anandasadagopan S. K., Karuppanan P., Venkatesan S., Sardana H. K., Kothari A., Jain R., Thakur A., Parihar D. S., Saifi A., Kaur J., Kumar V., Mishra A., Gogeri I., Rayasam G., Singh P., Chakraborty R., Chaturvedi G., Karunakar P., Yadav R., Singhmar S., Singh D., Sarkar S., Bhattacharya P., Acharya S., Singh V., Verma S., Soni D., Seth S., Vashisht S., Thakran S., Fatima F., Singh A. P., Sharma A., Sharma B., Subramanian M., Padwad Y. S., Hallan V., Patial V., Singh D., Tripude N. V., Chakrabarti P., Maity S. K., Ganguly D., Sarkar J., Ramakrishna S., Kumar B. N., Kumar K. A., Gandhi S. G., Jamwal P. S., Chouhan R., Jamwal V. L., Kapoor N., Ghosh D., Thakkar G., Subudhi U., Sen P., Chaudhury S. R., Kumar R., Gupta P., Tuli A., Sharma D., Ringe R. P., D A., Kulkarni M., Shanmugam D., Dharne M. S., Dastager S. G., Joshi R., Patil A. P., Mahajan S. N., Khan A. H., Wagh V., Yadav R. K., Khilari A., Bhadange M., Chaurasiya A. H., Kulsange S. E., Khairnar K., Paranjape S., Kalita J., Sastry N. G., Phukan T., Manna P., Romi W., Bharali P., Ozah D., Sahu R. K., Babu E. V., Sukumaran R., Nair A. R., Valappil P. K., Puthiyamadam A., Velayudhanpillai A., Chodankar K., Damare S., Madhavi Y., Aggarwal V. V., Dahiya S., Agrawal A., Dash D., Sengupta S., Insights from a Pan India Sero-Epidemiological survey (Phenome-India Cohort) for SARS-CoV2. eLife 10, e66537 (2021). 10.7554/eLife.66537 - DOI - PMC - PubMed
-
- Dan J. M., Mateus J., Kato Y., Hastie K. M., Yu E. D., Faliti C. E., Grifoni A., Ramirez S. I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S. A., Peters B., Krammer F., Simon V., Saphire E. O., Smith D. M., Weiskopf D., Sette A., Crotty S., Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371, eabf4063 (2021). 10.1126/science.abf4063 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

