Patterning with clocks and genetic cascades: Segmentation and regionalization of vertebrate versus insect body plans
- PMID: 34648490
- PMCID: PMC8516289
- DOI: 10.1371/journal.pgen.1009812
Patterning with clocks and genetic cascades: Segmentation and regionalization of vertebrate versus insect body plans
Abstract
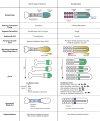
Oscillatory and sequential processes have been implicated in the spatial patterning of many embryonic tissues. For example, molecular clocks delimit segmental boundaries in vertebrates and insects and mediate lateral root formation in plants, whereas sequential gene activities are involved in the specification of regional identities of insect neuroblasts, vertebrate neural tube, vertebrate limb, and insect and vertebrate body axes. These processes take place in various tissues and organisms, and, hence, raise the question of what common themes and strategies they share. In this article, we review 2 processes that rely on the spatial regulation of periodic and sequential gene activities: segmentation and regionalization of the anterior-posterior (AP) axis of animal body plans. We study these processes in species that belong to 2 different phyla: vertebrates and insects. By contrasting 2 different processes (segmentation and regionalization) in species that belong to 2 distantly related phyla (arthropods and vertebrates), we elucidate the deep logic of patterning by oscillatory and sequential gene activities. Furthermore, in some of these organisms (e.g., the fruit fly Drosophila), a mode of AP patterning has evolved that seems not to overtly rely on oscillations or sequential gene activities, providing an opportunity to study the evolution of pattern formation mechanisms.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

