Genetic diversity of Salmonella Paratyphi A isolated from enteric fever patients in Bangladesh from 2008 to 2018
- PMID: 34648506
- PMCID: PMC8516307
- DOI: 10.1371/journal.pntd.0009748
Genetic diversity of Salmonella Paratyphi A isolated from enteric fever patients in Bangladesh from 2008 to 2018
Abstract
Background: The proportion of enteric fever cases caused by Salmonella Paratyphi A is increasing and may increase further as we begin to introduce typhoid conjugate vaccines (TCVs). While numerous epidemiological and genomic studies have been conducted for S. Typhi, there are limited data describing the genomic epidemiology of S. Paratyphi A in especially in endemic settings, such as Bangladesh.
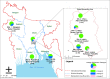
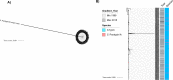
Principal findings: We conducted whole genome sequencing (WGS) of 67 S. Paratyphi A isolated between 2008 and 2018 from eight enteric disease surveillance sites across Bangladesh. We performed a detailed phylogenetic analysis of these sequence data incorporating sequences from 242 previously sequenced S. Paratyphi A isolates from a global collection and provided evidence of lineage migration from neighboring countries in South Asia. The data revealed that the majority of the Bangladeshi S. Paratyphi A isolates belonged to the dominant global lineage A (67.2%), while the remainder were either lineage C (19.4%) or F (13.4%). The population structure was relatively homogenous across the country as we did not find any significant lineage distributions between study sites inside or outside Dhaka. Our genomic data showed presence of single point mutations in gyrA gene either at codon 83 or 87 associated with decreased fluoroquinolone susceptibility in all Bangladeshi S. Paratyphi A isolates. Notably, we identified the pHCM2- like cryptic plasmid which was highly similar to S. Typhi plasmids circulating in Bangladesh and has not been previously identified in S. Paratyphi A organisms.
Significance: This study demonstrates the utility of WGS to monitor the ongoing evolution of this emerging enteric pathogen. Novel insights into the genetic structure of S. Paratyphi A will aid the understanding of both regional and global circulation patterns of this emerging pathogen and provide a framework for future genomic surveillance studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Arndt MB, Mosites EM, Tian M, Forouzanfar MH, Mokhdad AH, Meller M, et al.. Estimating the burden of paratyphoid a in Asia and Africa. PLoS neglected tropical diseases. 2014;8(6):e2925. Epub 2014/06/06. doi: 10.1371/journal.pntd.0002925 ; PubMed Central PMCID: PMC4046978 by Sanofi Pasteur, a commercial company, as the lead for Dengue Epidemiological studies. This does not alter our adherence to all PLOS NTDs policies on sharing data and materials. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

