Complete Genome Sequence of Weissella confusa LM1 and Comparative Genomic Analysis
- PMID: 34650545
- PMCID: PMC8506157
- DOI: 10.3389/fmicb.2021.749218
Complete Genome Sequence of Weissella confusa LM1 and Comparative Genomic Analysis
Abstract
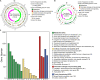
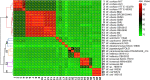
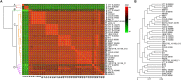
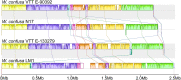
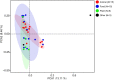
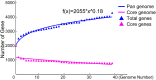
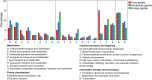
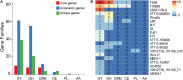
The genus Weissella is attracting an increasing amount of attention because of its multiple functions and probiotic potential. In particular, the species Weissella confusa is known to have great potential in industrial applications and exhibits numerous biological functions. However, the knowledge on this bacterium in insects is not investigated. Here, we isolated and identified W. confusa as the dominant lactic acid bacteria in the gut of the migratory locust. We named this strain W. confusa LM1, which is the first genome of an insect-derived W. confusa strain with one complete chromosome and one complete plasmid. Among all W. confusa strains, W. confusa LM1 had the largest genome. Its genome was the closest to that of W. confusa 1001271B_151109_G12, a strain from human feces. Our results provided accurate evolutionary relationships of known Weissella species and W. confusa strains. Based on genomic analysis, the pan-genome of W. confusa is in an open state. Most strains of W. confusa had the unique genes, indicating that these strains can adapt to different ecological niches and organisms. However, the variation of strain-specific genes did represent significant correlations with their hosts and ecological niches. These strains were predicted to have low potential to produce secondary metabolites. Furthermore, no antibiotic resistance genes were identified. At the same time, virulence factors associated with toxin production and secretion system were not found, indicating that W. confusa strains were not sufficient to perform virulence. Our study facilitated the discovery of the functions of W. confusa LM1 in locust biology and their potential application to locust management.
Keywords: Locusta migratoria; Weissella confusa; comparative genomics; gut bacteria; pan-genome.
Copyright © 2021 Yuan, Wang, Zhao and Kang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

