Associated SNPs, Heritabilities, Trait Correlations, and Genomic Breeding Values for Resistance in Snap Beans (Phaseolus vulgaris L.) to Root Rot Caused by Fusarium solani (Mart.) f. sp. phaseoli (Burkholder)
- PMID: 34650574
- PMCID: PMC8507974
- DOI: 10.3389/fpls.2021.697615
Associated SNPs, Heritabilities, Trait Correlations, and Genomic Breeding Values for Resistance in Snap Beans (Phaseolus vulgaris L.) to Root Rot Caused by Fusarium solani (Mart.) f. sp. phaseoli (Burkholder)
Abstract
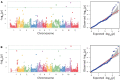
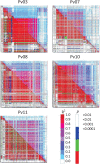
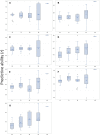
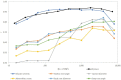
Root rot is a major constraint to snap bean (Phaseolus vulgaris) production in the United States and around the world. Genetic resistance is needed to effectively control root rot disease because cultural control methods are ineffective, and the pathogen will be present at the end of one season of production on previously clean land. A diversity panel of 149 snap bean pure lines was evaluated for resistance to Fusarium root rot in Oregon. Morphological traits potentially associated with root rot resistance, such as aboveground biomass, adventitious roots, taproot diameter, basal root diameter, deepest root angle, shallowest root angle, root angle average, root angle difference, and root angle geometric mean were evaluated and correlated to disease severity. A genome wide association study (GWAS) using the Fixed and random model Circulating Probability Unification (FarmCPU) statistical method, identified five associated single nucleotide polymorphisms (SNPs) for disease severity and two SNPs for biomass. The SNPs were found on Pv03, Pv07, Pv08, Pv10, and Pv11. One candidate gene for disease reaction near a SNP on Pv03 codes for a peroxidase, and two candidates associated with biomass SNPs were a 2-alkenal reductase gene cluster on Pv10 and a Pentatricopeptide repeat domain on Pv11. Bean lines utilized in the study were ranked by genomic estimated breeding values (GEBV) for disease severity, biomass, and the root architecture traits, and the observed and predicted values had high to moderate correlations. Cross validation of genomic predictions showed slightly lower correlational accuracy. Bean lines with the highest GEBV were among the most resistant, but did not necessarily rank at the very top numerically. This study provides information on the relationship of root architecture traits to root rot disease reaction. Snap bean lines with genetic merit for genomic selection were identified and may be utilized in future breeding efforts.
Keywords: best linear unbiased prediction; common bean; disease resistance; genome wide association studies; genomic prediction; genomic selection; root morphology.
Copyright © 2021 Huster, Wallace and Myers.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Genome-Wide Association Study Reveals Genomic Regions Associated with Fusarium Wilt Resistance in Common Bean.Genes (Basel). 2021 May 18;12(5):765. doi: 10.3390/genes12050765. Genes (Basel). 2021. PMID: 34069884 Free PMC article.
-
Genome-Wide Association Study and Genomic Prediction of Fusarium Wilt Resistance in Common Bean Core Collection.Int J Mol Sci. 2023 Oct 18;24(20):15300. doi: 10.3390/ijms242015300. Int J Mol Sci. 2023. PMID: 37894980 Free PMC article.
-
Screening for resistance to four fungal diseases and associated genomic regions in a snap bean diversity panel.Front Plant Sci. 2024 Jun 11;15:1386877. doi: 10.3389/fpls.2024.1386877. eCollection 2024. Front Plant Sci. 2024. PMID: 38919821 Free PMC article.
-
Genome-Wide Association Study and Genomic Prediction for Bacterial Wilt Resistance in Common Bean (Phaseolus vulgaris) Core Collection.Front Genet. 2022 May 31;13:853114. doi: 10.3389/fgene.2022.853114. eCollection 2022. Front Genet. 2022. PMID: 35711938 Free PMC article.
-
Characterization of fungal pathogens and germplasm screening for disease resistance in the main production area of the common bean in Argentina.Front Plant Sci. 2022 Sep 7;13:986247. doi: 10.3389/fpls.2022.986247. eCollection 2022. Front Plant Sci. 2022. PMID: 36161011 Free PMC article. Review.
Cited by
-
Horticultural performance and QTL mapping of snap bean (Phaseolus vulgaris L.) populations with organic and conventional breeding histories.Front Plant Sci. 2025 May 19;16:1533039. doi: 10.3389/fpls.2025.1533039. eCollection 2025. Front Plant Sci. 2025. PMID: 40538880 Free PMC article.
-
Influence of organic plant breeding on the rhizosphere microbiome of common bean (Phaseolus vulgaris L.).Front Plant Sci. 2023 Oct 25;14:1251919. doi: 10.3389/fpls.2023.1251919. eCollection 2023. Front Plant Sci. 2023. PMID: 37954997 Free PMC article.
-
A robust SNP-haplotype assay for Bct gene region conferring resistance to beet curly top virus in common bean (Phaseolus vulgaris L.).Front Plant Sci. 2023 Jul 14;14:1215950. doi: 10.3389/fpls.2023.1215950. eCollection 2023. Front Plant Sci. 2023. PMID: 37521933 Free PMC article.
References
-
- Abawi G., Crosier D., Cobb A. (1985). Root rot of snap beans in New York. New York’s Food and Life Sciences Bulletin. Geneva, NY: New York State Agricultural Experiment Station.
-
- Abawi G. S. (1990). Root rots of beans in Latin America and Africa: Diagnosis, research methodologies, and management strategies. Cali, Colombia: CIAT.
-
- Arenas S., Cortés A. J., Mastretta-Yanes A., Jaramillo-Correa J. P. (2021). Evaluating the accuracy of genomic prediction for the management and conservation of relictual natural tree populations. Tree Genet. Genom. 17 1–19.
-
- Assefa T., Mahama A. A., Brown A. V., Cannon E. K., Rubyogo J. C., Rao I. M., et al. (2019). A review of breeding objectives, genomic resources, and marker-assisted methods in common bean (Phaseolus vulgaris L.). Mol. Breed. 39:20. 10.1007/s11032-018-0920-0 - DOI
-
- Azzam H. A. (1956). Inheritance of resistance to Fusarium root rot in Phaseolus vulgaris L. and Phaseolus coccineus L. Ph.D. dissertation. Corvallis, OR: Oregon State University.
LinkOut - more resources
Full Text Sources

