Conditional and tissue-specific approaches to dissect essential mechanisms in plant development
- PMID: 34653951
- PMCID: PMC7612331
- DOI: 10.1016/j.pbi.2021.102119
Conditional and tissue-specific approaches to dissect essential mechanisms in plant development
Abstract
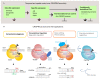
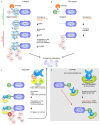
Reverse genetics approaches are routinely used to investigate gene function. However, mutations, especially in critical genes, can lead to pleiotropic effects as severe as lethality, thus limiting functional studies in specific contexts. Approaches that allow for modifications of genes or gene products in a specific spatial or temporal setting can overcome these limitations. The advent of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technologies has not only revolutionized targeted genome modification in plants but also enabled new possibilities for inducible and tissue-specific manipulation of gene functions at the DNA and RNA levels. In addition, novel approaches for the direct manipulation of target proteins have been introduced in plant systems. Here, we review the current development in tissue-specific and conditional manipulation approaches at the DNA, RNA, and protein levels.
Keywords: CRISPR; CRISPR-Activation; Conditional; Degron; Gene silencing; Nanobodies; Tissue-specific.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

