Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements
- PMID: 34654055
- PMCID: PMC9006278
- DOI: 10.1182/bloodadvances.2021006034
Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements
Abstract
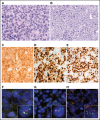
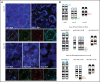
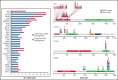
Diffuse large B-cell lymphoma (DLBCL) with aberrant coexpression of CD10+BCL6+MUM1+ (DLBCL-AE), classified as germinal center B cell (GCB) type by the Hans algorithm (HA), was genetically characterized. To capture the complexity of DLBCL-AE, we used an integrated approach that included gene expression profiling (GEP), fluorescence in situ hybridization, targeted gene sequencing, and copy number (CN) arrays. According to GEP, 32/54 (59%) cases were classified as GCB-DLBCL, 16/54 (30%) as activated B-cell (ABC) DLBCL, and 6/54 (11%) as unclassifiable. The discrepancy between HA and GEP was 41%. Three genetic subgroups were identified. Group 1 included 13/50 (26%) cases without translocations and mainly showing and ABC/MCD molecular profile. Group 2 comprised 11/50 (22%) cases with IRF4 alterations (DLBCL-IRF4), frequent mutations in IRF4 (82%) and NF-κB pathway genes (MYD88, CARD11, and CD79B), and losses of 17p13.2. Five cases each were classified as GCB- or ABC-type. Group 3 included 26/50 (52%) cases with 1 or several translocations in BCL2/BCL6/MYC/IGH, and GCB/EZB molecular profile predominated. Two cases in this latter group showed complex BCL2/BCL6/IRF4 translocations. DLBCL-IRF4 in adults showed a similar copy number profile and shared recurrent CARD11 and CD79B mutations when compared with LBCL-IRF4 in the pediatric population. However, adult cases showed higher genetic complexity, higher mutational load with frequent MYD88 and KMT2D mutations, and more ABC GEP. IRF4 mutations were identified only in IRF4-rearranged cases, indicating its potential use in the diagnostic setting. In conclusion, DLBCL-AE is genetically heterogeneous and enriched in cases with IRF4 alterations. DLBCL-IRF4 in adults has many similarities to the pediatric counterpart.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures




References
-
- Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015; 125(1):22-32. - PubMed
-
- Alizadeh AA, Eisen MB, Davis RE, et al. . Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000; 403(6769):503-511. - PubMed

