Comparative analysis of full-length mitochondrial genomes of five Skeletonema species reveals conserved genome organization and recent speciation
- PMID: 34654361
- PMCID: PMC8520197
- DOI: 10.1186/s12864-021-07999-z
Comparative analysis of full-length mitochondrial genomes of five Skeletonema species reveals conserved genome organization and recent speciation
Abstract
Background: Skeletonema species are prominent primary producers, some of which can also cause massive harmful algal blooms (HABs) in coastal waters under specific environmental conditions. Nevertheless, genomic information of Skeletonema species is currently limited, hindering advanced research on their role as primary producers and as HAB species. Mitochondrial genome (mtDNA) has been extensively used as "super barcode" in the phylogenetic analyses and comparative genomic analyses. However, of the 21 accepted Skeletonema species, full-length mtDNAs are currently available only for a single species, S. marinoi.
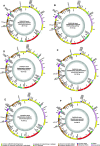
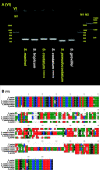
Results: In this study, we constructed full-length mtDNAs for six strains of five Skeletonema species, including S. marinoi, S. tropicum, S. grevillei, S. pseudocostatum and S. costatum (with two strains), which were isolated from coastal waters in China. The mtDNAs of all of these Skeletonema species were compact with short intergenic regions, no introns, and no repeat regions. Comparative analyses of these Skeletonema mtDNAs revealed high conservation, with a few discrete regions of high variations, some of which could be used as molecular markers for distinguishing Skeletonema species and for tracking the biogeographic distribution of these species with high resolution and specificity. We estimated divergence times among these Skeletonema species using 34 mtDNAs genes with fossil data as calibration point in PAML, which revealed that the Skeletonema species formed the independent clade diverging from Thalassiosira species approximately 48.30 Mya.
Conclusions: The availability of mtDNAs of five Skeletonema species provided valuable reference sequences for further evolutionary studies including speciation time estimation and comparative genomic analysis among diatom species. Divergent regions could be used as molecular markers for tracking different Skeletonema species in the fields of coastal regions.
Keywords: Comparative genomics; Divergence time; Harmful algal blooms; Mitochondrial genome; Molecular marker; Skeletonema species.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Sarno D, Kooistra WHCF, Medlin LK, Percopo I, Zingone A. Diversity in the genus skeletonema (bacillariophyceae). II. An assessment of the taxonomy of s. costatum-like species with the description of four new species. J Phycol. 2005;41(1):151–176. doi: 10.1111/j.1529-8817.2005.04067.x. - DOI
-
- M.D. Guiry in Guiry MDG, G.M . National University of Ireland. Galway: World-wide electronic publication; 2021. AlgaeBase.
-
- Xu X, Yu ZM, Cheng FJ, He LY, Cao XH, Song XX. Molecular diversity and ecological characteristics of the eukaryotic phytoplankton community in the coastal waters of the Bohai Sea, China. Harmful Algae. 2017;61:13–22. doi: 10.1016/j.hal.2016.11.005. - DOI
-
- Liu S, Chen N. Advances in biodiversity analysis of phytoplankton and harmful algal bloom species in the Jiaozhou Bay (Chinese with English abstract). Mar Sci. 2021;45(4):170–88.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

