Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology
- PMID: 34654800
- PMCID: PMC8520019
- DOI: 10.1038/s41467-021-25942-4
Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology
Erratum in
-
Publisher Correction: Compensatory ion transport buffers daily protein rhythms to regulate osmotic balance and cellular physiology.Nat Commun. 2021 Nov 24;12(1):6988. doi: 10.1038/s41467-021-26725-7. Nat Commun. 2021. PMID: 34819501 Free PMC article. No abstract available.
Abstract
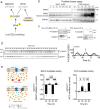
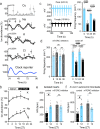
Between 6-20% of the cellular proteome is under circadian control and tunes mammalian cell function with daily environmental cycles. For cell viability, and to maintain volume within narrow limits, the daily variation in osmotic potential exerted by changes in the soluble proteome must be counterbalanced. The mechanisms and consequences of this osmotic compensation have not been investigated before. In cultured cells and in tissue we find that compensation involves electroneutral active transport of Na+, K+, and Cl- through differential activity of SLC12A family cotransporters. In cardiomyocytes ex vivo and in vivo, compensatory ion fluxes confer daily variation in electrical activity. Perturbation of soluble protein abundance has commensurate effects on ion composition and cellular function across the circadian cycle. Thus, circadian regulation of the proteome impacts ion homeostasis with substantial consequences for the physiology of electrically active cells such as cardiomyocytes.
© 2021. Crown.
Conflict of interest statement
P.N. holds shares in AstraZeneca. The other authors declare no competing interests.
Figures


References
-
- Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr. Biol. 2006;16:1107–1115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

