Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway
- PMID: 34654815
- PMCID: PMC8521590
- DOI: 10.1038/s41467-021-26330-8
Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway
Abstract
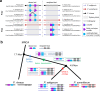
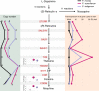
For millions of years, plants evolve plenty of structurally diverse secondary metabolites (SM) to support their sessile lifestyles through continuous biochemical pathway innovation. While new genes commonly drive the evolution of plant SM pathway, how a full biosynthetic pathway evolves remains poorly understood. The evolution of pathway involves recruiting new genes along the reaction cascade forwardly, backwardly, or in a patchwork manner. With three chromosome-scale Papaver genome assemblies, we here reveal whole-genome duplications (WGDs) apparently accelerate chromosomal rearrangements with a nonrandom distribution towards SM optimization. A burst of structural variants involving fusions, translocations and duplications within 7.7 million years have assembled nine genes into the benzylisoquinoline alkaloids gene cluster, following a punctuated patchwork model. Biosynthetic gene copies and their total expression matter to morphinan production. Our results demonstrate how new genes have been recruited from a WGD-induced repertoire of unregulated enzymes with promiscuous reactivities to innovate efficient metabolic pathways with spatiotemporal constraint.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Reply to "Subgenome-aware analyses suggest a reticulate allopolyploidization origin in three Papaver genomes".Nat Commun. 2023 Apr 19;14(1):2203. doi: 10.1038/s41467-023-37940-9. Nat Commun. 2023. PMID: 37076521 Free PMC article. No abstract available.
-
Subgenome-aware analyses suggest a reticulate allopolyploidization origin in three Papaver genomes.Nat Commun. 2023 Apr 19;14(1):2204. doi: 10.1038/s41467-023-37939-2. Nat Commun. 2023. PMID: 37076529 Free PMC article. No abstract available.
Similar articles
-
The opium poppy genome and morphinan production.Science. 2018 Oct 19;362(6412):343-347. doi: 10.1126/science.aat4096. Epub 2018 Aug 30. Science. 2018. PMID: 30166436
-
Benzylisoquinoline alkaloid biosynthesis in opium poppy.Planta. 2014 Jul;240(1):19-32. doi: 10.1007/s00425-014-2056-8. Epub 2014 Mar 27. Planta. 2014. PMID: 24671624 Review.
-
Over 100 Million Years of Enzyme Evolution Underpinning the Production of Morphine in the Papaveraceae Family of Flowering Plants.Plant Commun. 2020 Mar 9;1(2):100029. doi: 10.1016/j.xplc.2020.100029. Plant Commun. 2020. PMID: 32685922 Free PMC article.
-
A functionally conserved STORR gene fusion in Papaver species that diverged 16.8 million years ago.Nat Commun. 2022 Jun 7;13(1):3150. doi: 10.1038/s41467-022-30856-w. Nat Commun. 2022. PMID: 35672295 Free PMC article.
-
Evolution of morphine biosynthesis in opium poppy.Phytochemistry. 2009 Oct-Nov;70(15-16):1696-707. doi: 10.1016/j.phytochem.2009.07.006. Epub 2009 Aug 6. Phytochemistry. 2009. PMID: 19665152 Review.
Cited by
-
A cornucopia of diversity-Ranunculales as a model lineage.J Exp Bot. 2024 Mar 27;75(7):1800-1822. doi: 10.1093/jxb/erad492. J Exp Bot. 2024. PMID: 38109712 Free PMC article. Review.
-
TCMPG: an integrative database for traditional Chinese medicine plant genomes.Hortic Res. 2022 Mar 8;9:uhac060. doi: 10.1093/hr/uhac060. eCollection 2022. Hortic Res. 2022. PMID: 35591924 Free PMC article.
-
The tissue-specific chromatin accessibility landscape of Papaver somniferum.Front Genet. 2023 Mar 15;14:1136736. doi: 10.3389/fgene.2023.1136736. eCollection 2023. Front Genet. 2023. PMID: 37007951 Free PMC article. No abstract available.
-
SOI: robust identification of orthologous synteny with the Orthology Index and broad applications in evolutionary genomics.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf320. doi: 10.1093/nar/gkaf320. Nucleic Acids Res. 2025. PMID: 40248914 Free PMC article.
-
Evolution and diversification of the momilactone biosynthetic gene cluster in the genus Oryza.New Phytol. 2025 Mar;245(6):2681-2697. doi: 10.1111/nph.20416. Epub 2025 Jan 30. New Phytol. 2025. PMID: 39887739 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

