Genome instability independent of type I interferon signaling drives neuropathology caused by impaired ribonucleotide excision repair
- PMID: 34655526
- PMCID: PMC8686690
- DOI: 10.1016/j.neuron.2021.09.040
Genome instability independent of type I interferon signaling drives neuropathology caused by impaired ribonucleotide excision repair
Abstract
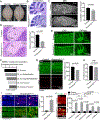
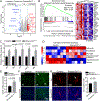
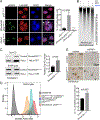
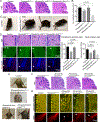
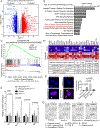
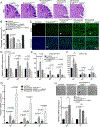
Aicardi-Goutières syndrome (AGS) is a monogenic type I interferonopathy characterized by neurodevelopmental defects and upregulation of type I interferon signaling and neuroinflammation. Mutations in genes that function in nucleic acid metabolism, including RNASEH2, are linked to AGS. Ribonuclease H2 (RNASEH2) is a genome surveillance factor critical for DNA integrity by removing ribonucleotides incorporated into replicating DNA. Here we show that RNASEH2 is necessary for neurogenesis and to avoid activation of interferon-responsive genes and neuroinflammation. Cerebellar defects after RNASEH2B inactivation are rescued by p53 but not cGAS deletion, suggesting that DNA damage signaling, not neuroinflammation, accounts for neuropathology. Coincident inactivation of Atm and Rnaseh2 further affected cerebellar development causing ataxia, which was dependent upon aberrant activation of non-homologous end-joining (NHEJ). The loss of ATM also markedly exacerbates cGAS-dependent type I interferon signaling. Thus, DNA damage-dependent signaling rather than type I interferon signaling underlies neurodegeneration in this class of neurodevelopmental/neuroinflammatory disease.
Keywords: ATM; Aicardi-Goutières syndrome; Cerebellum; DNA damage; Microglia; Neurodegeneration; Neurodevelopment; Neuroinflammation; RNaseH2; cGAS/STING.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Comment in
-
DNA damage rather than type I IFN signaling is the primary mediator of neural dysfunction in Aicardi-Goutières syndrome after RNASEH2 disruption.Neuron. 2021 Dec 15;109(24):3897-3900. doi: 10.1016/j.neuron.2021.11.019. Neuron. 2021. PMID: 34914915
References
-
- Abdel-Salam GM, Zaki MS, Lebon P, and Meguid NA (2004). Aicardi-Goutieres syndrome: clinical and neuroradiological findings of 10 new cases. Acta Paediatr 93, 929–936. - PubMed
-
- Aicardi J, and Goutieres F (1984). A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol 15, 49–54. - PubMed
-
- Akwa Y, Hassett DE, Eloranta ML, Sandberg K, Masliah E, Powell H, Whitton JL, Bloom FE, and Campbell IL (1998). Transgenic expression of IFN-alpha in the central nervous system of mice protects against lethal neurotropic viral infection but induces inflammation and neurodegeneration. J Immunol 161, 5016–5026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

