ClineHelpR: an R package for genomic cline outlier detection and visualization
- PMID: 34656096
- PMCID: PMC8520269
- DOI: 10.1186/s12859-021-04423-x
ClineHelpR: an R package for genomic cline outlier detection and visualization
Abstract
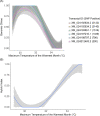
Background: Patterns of multi-locus differentiation (i.e., genomic clines) often extend broadly across hybrid zones and their quantification can help diagnose how species boundaries are shaped by adaptive processes, both intrinsic and extrinsic. In this sense, the transitioning of loci across admixed individuals can be contrasted as a function of the genome-wide trend, in turn allowing an expansion of clinal theory across a much wider array of biodiversity. However, computational tools that serve to interpret and consequently visualize 'genomic clines' are limited, and users must often write custom, relatively complex code to do so.
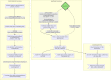
Results: Here, we introduce the ClineHelpR R-package for visualizing genomic clines and detecting outlier loci using output generated by two popular software packages, bgc and Introgress. ClineHelpR bundles both input generation (i.e., filtering datasets and creating specialized file formats) and output processing (e.g., MCMC thinning and burn-in) with functions that directly facilitate interpretation and hypothesis testing. Tools are also provided for post-hoc analyses that interface with external packages such as ENMeval and RIdeogram.
Conclusions: Our package increases the reproducibility and accessibility of genomic cline methods, thus allowing an expanded user base and promoting these methods as mechanisms to address diverse evolutionary questions in both model and non-model organisms. Furthermore, the ClineHelpR extended functionality can evaluate genomic clines in the context of spatial and environmental features, allowing users to explore underlying processes potentially contributing to the observed patterns and helping facilitate effective conservation management strategies.
Keywords: Genomic cline; Hybrid zones; Introgression; Outlier detection; Population genetics; Selection; bgc.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

