A novel phosphoproteomic landscape evoked in response to type I interferon in the brain and in glial cells
- PMID: 34656141
- PMCID: PMC8520650
- DOI: 10.1186/s12974-021-02277-x
A novel phosphoproteomic landscape evoked in response to type I interferon in the brain and in glial cells
Abstract
Background: Type I interferons (IFN-I) are key responders to central nervous system infection and injury and are also increased in common neurodegenerative diseases. Their effects are primarily mediated via transcriptional regulation of several hundred interferon-regulated genes. In addition, IFN-I activate several kinases including members of the MAPK and PI3K families. Yet, how changes to the global protein phosphoproteome contribute to the cellular response to IFN-I is unknown.
Methods: The cerebral phosphoproteome of mice with brain-targeted chronic production of the IFN-I, IFN-α, was obtained. Changes in phosphorylation were analyzed by ontology and pathway analysis and kinase enrichment predictions. These were verified by phenotypic analysis, immunohistochemistry and immunoblots. In addition, primary murine microglia and astrocytes, the brain's primary IFN-I-responding cells, were acutely treated with IFN-α and the global phosphoproteome was similarly analyzed.
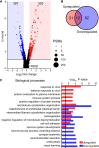
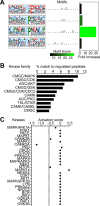
Results: We identified widespread protein phosphorylation as a novel mechanism by which IFN-I mediate their effects. In our mouse model for IFN-I-induced neurodegeneration, protein phosphorylation, rather than the proteome, aligned with the clinical hallmarks and pathological outcome, including impaired development, motor dysfunction and seizures. In vitro experiments revealed extensive and rapid IFN-I-induced protein phosphorylation in microglia and astrocytes. Response to acute IFN-I stimulation was independent of gene expression and mediated by a small number of kinase families. The changes in the phosphoproteome affected a diverse range of cellular processes and functional analysis suggested that this response induced an immediate reactive state and prepared cells for subsequent transcriptional responses.
Conclusions: Our studies reveal a hitherto unappreciated role for changes in the protein phosphorylation landscape in cellular responses to IFN-I and thus provide insights for novel diagnostic and therapeutic strategies for neurological diseases caused by IFN-I.
Keywords: Astrocyte; Cerebral type I interferonopathy; Interferon; Microglia; Neurodegenerative disease; Phosphoproteomics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

