Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana
- PMID: 34657568
- PMCID: PMC9225624
- DOI: 10.1080/15548627.2021.1976965
Mechanistic insights into an atypical interaction between ATG8 and SH3P2 in Arabidopsis thaliana
Abstract
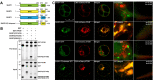
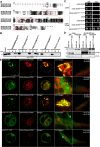
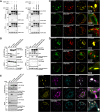
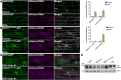
In selective macroautophagy/autophagy, cargo receptors are recruited to the forming autophagosome by interacting with Atg8 (autophagy-related 8)-family proteins and facilitate the selective sequestration of specific cargoes for autophagic degradation. In addition, Atg8 interacts with a number of adaptors essential for autophagosome biogenesis, including ATG and non-ATG proteins. The majority of these adaptors and receptors are characterized by an Atg8-family interacting motif (AIM) for binding to Atg8. However, the molecular basis for the interaction mode between ATG8 and regulators or cargo receptors in plants remains largely unclear. In this study, we unveiled an atypical interaction mode for Arabidopsis ATG8f with a plant unique adaptor protein, SH3P2 (SH3 domain-containing protein 2), but not with the other two SH3 proteins. By structure analysis of the unbound form of ATG8f, we identified the unique conformational changes in ATG8f upon binding to the AIM sequence of a plant known autophagic receptor, NBR1. To compare the binding affinity of SH3P2-ATG8f with that of ATG8f-NBR1, we performed a gel filtration assay to show that ubiquitin-associated domain of NBR1 outcompetes the SH3 domain of SH3P2 for ATG8f interaction. Biochemical and cellular analysis revealed that distinct interfaces were employed by ATG8f to interact with NBR1 and SH3P2. Further subcellular analysis showed that the AIM-like motif of SH3P2 is essential for its recruitment to the phagophore membrane but is dispensable for its trafficking in endocytosis. Taken together, our study provides an insightful structural basis for the ATG8 binding specificity toward a plant-specific autophagic adaptor and a conserved autophagic receptor.Abbreviations: ATG, autophagy-related; AIM, Atg8-family interacting motif; BAR, Bin-Amphiphysin-Rvs; BFA, brefeldin A; BTH, benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester; CCV, clathrin-coated-vesicle; CLC2, clathrin light chain 2; Conc A, concanamycin A; ER, endoplasmic reticulum; LDS, LIR docking site; MAP1LC3/LC3, microtubule associated protein 1 light chain 3; LIR, LC3-interacting region; PE, phosphatidylethanolamine; SH3P2, SH3 domain containing protein 2; SH3, Src-Homology-3; UBA, ubiquitin-associated; UIM, ubiquitin-interacting motif.
Keywords: ATG8 interacting motif; Arabidopsis ATG8; NBR1; SH3P2; selective autophagy.
Figures


References
-
- Liu Y, Bassham DC.. Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol. 2012;63:215–237. - PubMed
-
- Zhuang X, Chung KP, Luo M, et al. Autophagosome biogenesis and the endoplasmic reticulum: a plant perspective. Trends Plant Sci. 2018;23(8):677–692. - PubMed
-
- Michaeli S, Galili G, Genschik P, et al. Autophagy in plants–What’s new on the menu? Trends Plant Sci. 2016;21(2):134–144. - PubMed
-
- Floyd BE, Morriss SC, MacIntosh GC, et al. What to eat: evidence for selective autophagy in plants. J Integr Plant Biol. 2012;54(11):907–920 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
