The OpenNeuro resource for sharing of neuroscience data
- PMID: 34658334
- PMCID: PMC8550750
- DOI: 10.7554/eLife.71774
The OpenNeuro resource for sharing of neuroscience data
Abstract
The sharing of research data is essential to ensure reproducibility and maximize the impact of public investments in scientific research. Here, we describe OpenNeuro, a BRAIN Initiative data archive that provides the ability to openly share data from a broad range of brain imaging data types following the FAIR principles for data sharing. We highlight the importance of the Brain Imaging Data Structure standard for enabling effective curation, sharing, and reuse of data. The archive presently shares more than 600 datasets including data from more than 20,000 participants, comprising multiple species and measurement modalities and a broad range of phenotypes. The impact of the shared data is evident in a growing number of published reuses, currently totalling more than 150 publications. We conclude by describing plans for future development and integration with other ongoing open science efforts.
Keywords: EEG; MEG; MRI; data sharing; human; mouse; neuroimaging; neuroscience; open science; rat.
© 2021, Markiewicz et al.
Conflict of interest statement
CM, KG, FF, RB, YH, JW, OE, MG, AJ, RP No competing interests declared, EM is owner of Squishymedia which is funded to perform software development work on OpenNeuro. NH is an employee of Squishymedia which is funded to perform software development work on OpenNeuro.
Figures



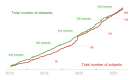

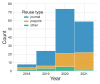
Comment in
-
A FAIR platform for data-sharing.Nat Neurosci. 2021 Dec;24(12):1640. doi: 10.1038/s41593-021-00976-5. Nat Neurosci. 2021. PMID: 34848877 No abstract available.
References
-
- Bannier E, Barker G, Borghesani V, Broeckx N, Clement P, Emblem KE, Ghosh S, Glerean E, Gorgolewski KJ, Havu M, Halchenko YO, Herholz P, Hespel A, Heunis S, Hu Y, Hu C-P, Huijser D, de la Iglesia Vayá M, Jancalek R, Katsaros VK, Kieseler M-L, Maumet C, Moreau CA, Mutsaerts H-J, Oostenveld R, Ozturk-Isik E, Pascual Leone Espinosa N, Pellman J, Pernet CR, Pizzini FB, Trbalić AŠ, Toussaint P-J, Visconti di Oleggio Castello M, Wang F, Wang C, Zhu H. The Open Brain Consent: Informing research participants and obtaining consent to share brain imaging data. Human Brain Mapping. 2021;42:1945–1951. doi: 10.1002/hbm.25351. - DOI - PMC - PubMed
-
- Bansal S, Kori A, Zulfikar W, Wexler J, Markiewicz C, Feingold F, Poldrack R. High-Sensitivity Detection of Facial Features on MRI Brain Scans with a Convolutional Network. bioRxiv. 2020 doi: 10.1101/2021.04.25.441373. - DOI
-
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kötter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts S, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP. Toward discovery science of human brain function. PNAS. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

