Enhanced Susceptibility of ADAP-Deficient Mice to Listeria monocytogenes Infection Is Associated With an Altered Phagocyte Phenotype and Function
- PMID: 34659211
- PMCID: PMC8515145
- DOI: 10.3389/fimmu.2021.724855
Enhanced Susceptibility of ADAP-Deficient Mice to Listeria monocytogenes Infection Is Associated With an Altered Phagocyte Phenotype and Function
Abstract
The adhesion and degranulation-promoting adaptor protein (ADAP) serves as a multifunctional scaffold and is involved in the formation of immune signaling complexes. To date, only limited data exist regarding the role of ADAP in pathogen-specific immunity during in vivo infection, and its contribution in phagocyte-mediated antibacterial immunity remains elusive. Here, we show that mice lacking ADAP (ADAPko) are highly susceptible to the infection with the intracellular pathogen Listeria monocytogenes (Lm) by showing enhanced immunopathology in infected tissues together with increased morbidity, mortality, and excessive infiltration of neutrophils and monocytes. Despite high phagocyte numbers in the spleen and liver, ADAPko mice only inefficiently controlled pathogen growth, hinting at a functional impairment of infection-primed phagocytes in the ADAP-deficient host. Flow cytometric analysis of hallmark pro-inflammatory mediators and unbiased whole genome transcriptional profiling of neutrophils and inflammatory monocytes uncovered broad molecular alterations in the inflammatory program in both phagocyte subsets following their activation in the ADAP-deficient host. Strikingly, ex vivo phagocytosis assay revealed impaired phagocytic capacity of neutrophils derived from Lm-infected ADAPko mice. Together, our data suggest that an alternative priming of phagocytes in ADAP-deficient mice during Lm infection induces marked alterations in the inflammatory profile of neutrophils and inflammatory monocytes that contribute to enhanced immunopathology while limiting their capacity to eliminate the pathogen and to prevent the fatal outcome of the infection.
Keywords: ADAP; Listeria monocytogenes; adaptor protein; inflammatory monocytes; neutrophil extracellular traps; neutrophils; phagocytosis.
Copyright © 2021 Böning, Parzmair, Jeron, Düsedau, Kershaw, Xu, Relja, Schlüter, Dunay, Reinhold, Schraven and Bruder.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
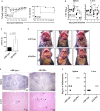




 > 3 beads,
> 3 beads,  3 beads,
3 beads,  2 beads,
2 beads,  1 bead, and
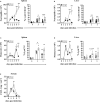
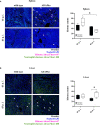
1 bead, and  0 beads) show the fractioned cells according to the amount of incorporated beads. Phagocytic capability of (B, left panel) spleen and (C, left panel) liver neutrophils was considered by the MFI of bead-positive populations. Data are depicted as mean ± SEM for n = 6–8 individually analyzed mice per group out of two independent experiments. (B, C, left panels) Statistical analyses were performed using two-way ANOVA with Bonferroni’s post-hoc test and (B, C, right panels) two-tailed unpaired t-test with Welch’s correction (**p < 0.01, ****p < 0.0001). ADAPko, adhesion and degranulation-promoting adaptor protein knockout; CFU, colony-forming units; FITC, fluorescein isothiocyanate; MFI, mean fluorescence intensity. *p < 0.05, ***p < 0.001.
0 beads) show the fractioned cells according to the amount of incorporated beads. Phagocytic capability of (B, left panel) spleen and (C, left panel) liver neutrophils was considered by the MFI of bead-positive populations. Data are depicted as mean ± SEM for n = 6–8 individually analyzed mice per group out of two independent experiments. (B, C, left panels) Statistical analyses were performed using two-way ANOVA with Bonferroni’s post-hoc test and (B, C, right panels) two-tailed unpaired t-test with Welch’s correction (**p < 0.01, ****p < 0.0001). ADAPko, adhesion and degranulation-promoting adaptor protein knockout; CFU, colony-forming units; FITC, fluorescein isothiocyanate; MFI, mean fluorescence intensity. *p < 0.05, ***p < 0.001.Similar articles
-
ADAP Promotes Degranulation and Migration of NK Cells Primed During in vivo Listeria monocytogenes Infection in Mice.Front Immunol. 2020 Jan 22;10:3144. doi: 10.3389/fimmu.2019.03144. eCollection 2019. Front Immunol. 2020. PMID: 32038647 Free PMC article.
-
IκBNS-deficiency protects mice from fatal Listeria monocytogenes infection by blunting pro-inflammatory signature in Ly6Chigh monocytes and preventing exaggerated innate immune responses.Front Immunol. 2022 Dec 22;13:1028789. doi: 10.3389/fimmu.2022.1028789. eCollection 2022. Front Immunol. 2022. PMID: 36618344 Free PMC article.
-
ADAP plays a pivotal role in CD4+ T cell activation but is only marginally involved in CD8+ T cell activation, differentiation, and immunity to pathogens.J Leukoc Biol. 2017 Feb;101(2):407-419. doi: 10.1189/jlb.1A0216-090RR. Epub 2016 Sep 7. J Leukoc Biol. 2017. PMID: 27605210
-
The Essential Role of Neutrophils during Infection with the Intracellular Bacterial Pathogen Listeria monocytogenes.J Immunol. 2016 Sep 1;197(5):1557-65. doi: 10.4049/jimmunol.1600599. J Immunol. 2016. PMID: 27543669 Free PMC article. Review.
-
Early host-pathogen interactions in the liver and spleen during systemic murine listeriosis: an overview.Immunobiology. 1999 Dec;201(2):178-87. doi: 10.1016/S0171-2985(99)80057-6. Immunobiology. 1999. PMID: 10631566 Review.
Cited by
-
FYB1-targeted modulation of CAPG promotes AML progression.Mol Cell Biochem. 2025 Feb;480(2):985-999. doi: 10.1007/s11010-024-04992-4. Epub 2024 May 3. Mol Cell Biochem. 2025. PMID: 38700746 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

