Transcriptome Profiling Reveals Important Transcription Factors and Biological Processes in Skin Regeneration Mediated by Mechanical Stretch
- PMID: 34659370
- PMCID: PMC8511326
- DOI: 10.3389/fgene.2021.757350
Transcriptome Profiling Reveals Important Transcription Factors and Biological Processes in Skin Regeneration Mediated by Mechanical Stretch
Abstract
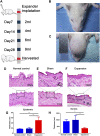
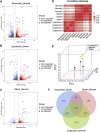
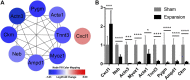
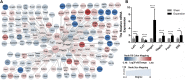
Background: Mechanical stretch is utilized to promote skin regeneration during tissue expansion for reconstructive surgery. Although mechanical stretch induces characteristic morphological changes in the skin, the biological processes and molecular mechanisms involved in mechanically induced skin regeneration are not well elucidated. Methods: A male rat scalp expansion model was established and the important biological processes related to mechanical stretch-induced skin regeneration were identified using Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and gene set enrichment analysis (GSEA). Analysis was also conducted by constructing a protein-protein interaction (PPI) network, identifying key modules and hub genes, determining transcription factor (TF)-mRNA regulatory relationships, and confirming the expression pattern of the TFs and hub genes. Results: We identified nine robust hub genes (CXCL1, NEB, ACTN3, MYOZ1, ACTA1, TNNT3, PYGM, AMPD1, and CKM) that may serve as key molecules in skin growth. These genes were determined to be involved in several important biological processes, including keratinocyte differentiation, cytoskeleton reorganization, chemokine signaling pathway, glycogen metabolism, and voltage-gated ion channel activity. The potentially significant pathways, including the glucagon signaling pathway, the Wnt signaling pathway, and cytokine-cytokine receptor interaction, were distinguished. In addition, we identified six TFs (LEF1, TCF7, HMGA1, TFAP2C, FOSL1, and ELF5) and constructed regulatory TF-mRNA interaction networks. Conclusion: This study generated a comprehensive overview of the gene networks underlying mechanically induced skin regeneration. The functions of these key genes and the pathways in which they participate may reveal new aspects of skin regeneration under mechanical strain. Furthermore, the identified TF regulators can be used as potential candidates for clinical therapeutics for skin pretreatment before reconstructive surgery.
Keywords: hub genes; mechanical stretch; skin regeneration; tissue expansion; transcription facotrs; transcriptome.
Copyright © 2021 Liu, Xiong, Zhang, Du, Dong, Yu and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

