MEMES: Machine learning framework for Enhanced MolEcular Screening
- PMID: 34659706
- PMCID: PMC8442698
- DOI: 10.1039/d1sc02783b
MEMES: Machine learning framework for Enhanced MolEcular Screening
Abstract
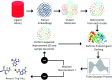
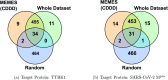
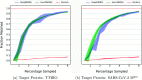
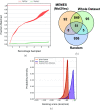
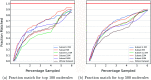
In drug discovery applications, high throughput virtual screening exercises are routinely performed to determine an initial set of candidate molecules referred to as "hits". In such an experiment, each molecule from a large small-molecule drug library is evaluated in terms of physical properties such as the docking score against a target receptor. In real-life drug discovery experiments, drug libraries are extremely large but still there is only a minor representation of the essentially infinite chemical space, and evaluation of physical properties for each molecule in the library is not computationally feasible. In the current study, a novel Machine learning framework for Enhanced MolEcular Screening (MEMES) based on Bayesian optimization is proposed for efficient sampling of the chemical space. The proposed framework is demonstrated to identify 90% of the top-1000 molecules from a molecular library of size about 100 million, while calculating the docking score only for about 6% of the complete library. We believe that such a framework would tremendously help to reduce the computational effort in not only drug-discovery but also areas that require such high-throughput experiments.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
International Institute of Information Technology, Hyderabad has filed provisional patent application for the use of the MEMES framework in high-throughput screening exercises, with U. D. P., S. M., S. L., and Y. P. listed as inventors. Provisional patent application No.: 202041050608. Application status: awaiting complete specification (provisional patent filed). The funders did not have any role in the design, idea, data collection, analysis, interpretation, writing of the manuscript or decision to submit it for publication.
Figures


References
LinkOut - more resources
Full Text Sources

