Exploration of the Role of the C-Terminal Domain of Human DNA Topoisomerase IIα in Catalytic Activity
- PMID: 34660952
- PMCID: PMC8515377
- DOI: 10.1021/acsomega.1c02083
Exploration of the Role of the C-Terminal Domain of Human DNA Topoisomerase IIα in Catalytic Activity
Abstract
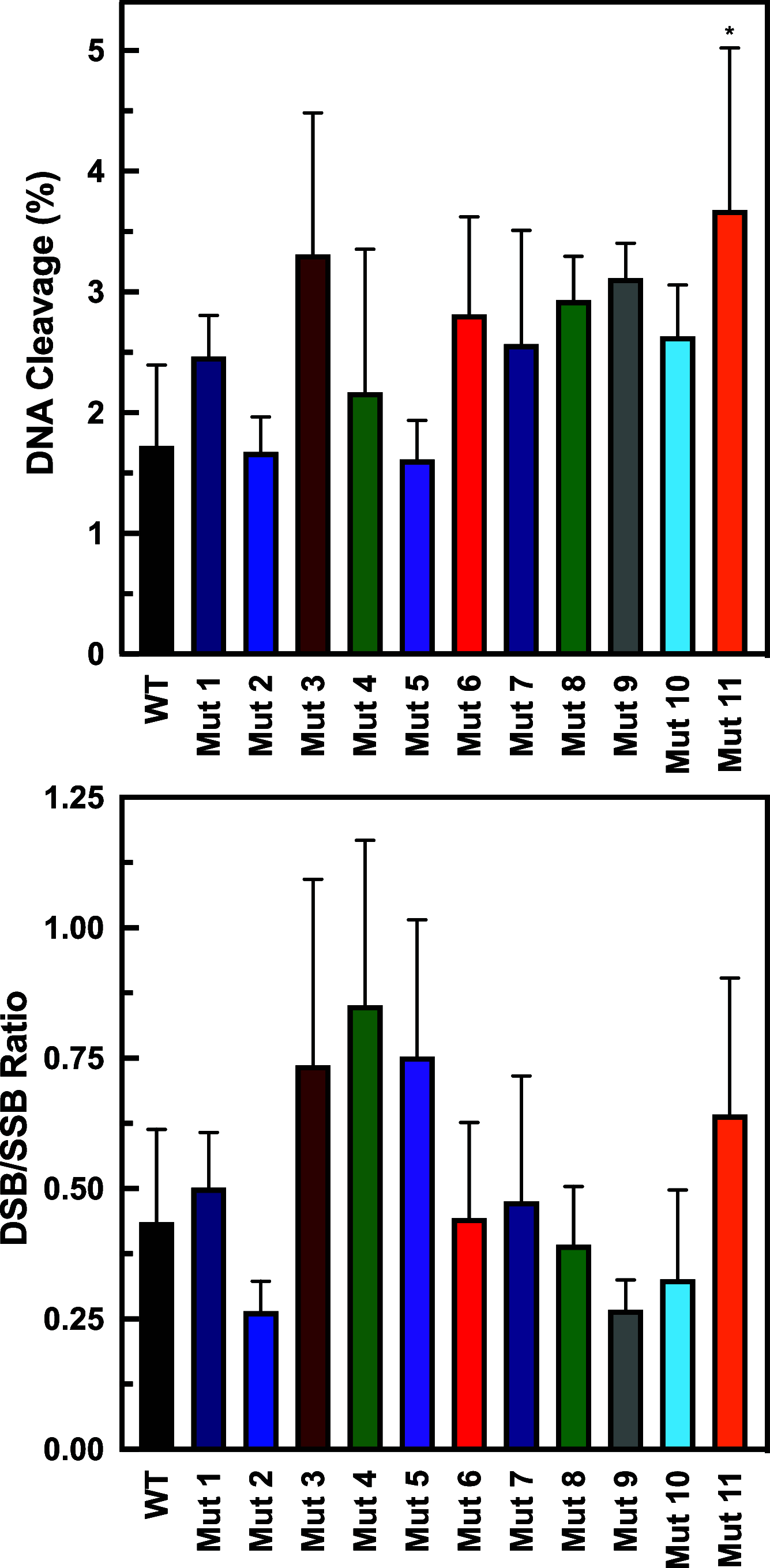
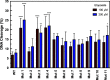
Human topoisomerase IIα (TOP2A) is a vital nuclear enzyme involved in resolving knots and tangles in DNA during replication and cell division. TOP2A is a homodimer with a symmetrical, multidomain structure. While the N-terminal and core regions of the protein are well-studied, the C-terminal domain is poorly understood but is involved in enzyme regulation and is predicted to be intrinsically disordered. In addition, it appears to be a major region of post-translational modification and includes several Ser and Thr residues, many of which have not been studied for biochemical effects. Therefore, we generated a series of human TOP2A mutants where we changed specific Ser and Thr residues in the C-terminal domain to Ala, Gly, or Ile residues. We designed, purified, and examined 11 mutant TOP2A enzymes. The amino acid changes were made between positions 1272 and 1525 with 1-7 residues changed per mutant. Several mutants displayed increased levels of DNA cleavage without displaying any change in plasmid DNA relaxation or DNA binding. For example, mutations in the regions 1272-1279, 1324-1343, 1351-1365, and 1374-1377 produced 2-3 times more DNA cleavage in the presence of etoposide than wild-type TOP2A. Further, several mutants displayed changes in relaxation and/or decatenation activity. Together, these results support previous findings that the C-terminal domain of TOP2A influences catalytic activity and interacts with the substrate DNA. Furthermore, we hypothesize that it may be possible to regulate the enzyme by targeting positions in the C-terminal domain. Because the C-terminal domain differs between the two human TOP2 isoforms, this strategy may provide a means for selectively targeting TOP2A for therapeutic inhibition. Additional studies are warranted to explore these results in more detail.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Murphy M. B.; Mercer S. L.; Deweese J. E.. Inhibitors and Poisons of Mammalian Type II Topoisomerases. In Advances in Molecular Toxicology; Fishbein J. C.; Heilman J., Eds.; Academic Press: Cambridge, MA, 2017; Vol. 11, pp 203–240.
LinkOut - more resources
Full Text Sources
Miscellaneous

