Visualized and Quantitative Conformational Analysis of Peptidomimetics
- PMID: 34661014
- PMCID: PMC8515614
- DOI: 10.1021/acsomega.1c03967
Visualized and Quantitative Conformational Analysis of Peptidomimetics
Abstract
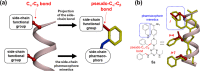
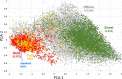
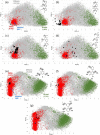
Protein-protein interactions (PPIs) are fundamentally important and challenging drug targets. Peptidomimetic molecules of various types have been developed to modulate PPIs. A particularly promising drug discovery strategy, structural peptidomimetics, was designed based on special mimicking of side-chain Cα-Cβ bonds. It is simple and versatile. Nevertheless, no quantitative method has been established to evaluate its similarity to a target peptide motif. We developed two methods that enable visual, comprehensive, and quantitative analysis of peptidomimetics: peptide conformation distribution (PCD) plot and peptidomimetic analysis (PMA) map. These methods specifically examine multiple side-chain Cα-Cβ bonds of a peptide fragment motif and their corresponding bonds (pseudo-Cα-Cβ bonds) in a mimetic molecule instead of φ and ψ angles of a single amino acid in the traditional Ramachandran plot. The PCD plot is an alignment-free method, whereas the PMA map is an alignment-based method providing distinctive and complementary analysis. Results obtained from analysis using these two methods indicate our multifacial α-helix mimetic scaffold 12 as an excellent peptidomimetic that can precisely mimic the spatial positioning of side-chain functional groups of α-helix. These methods are useful for visualized and quantified evaluation of peptidomimetics and for the rational design of new mimetic scaffolds.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
LinkOut - more resources
Full Text Sources

