A Probabilistic Programming Approach to Protein Structure Superposition
- PMID: 34661202
- PMCID: PMC8515897
- DOI: 10.1109/cibcb.2019.8791469
A Probabilistic Programming Approach to Protein Structure Superposition
Abstract
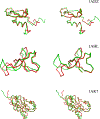
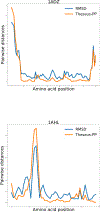
Optimal superposition of protein structures or other biological molecules is crucial for understanding their structure, function, dynamics and evolution. Here, we investigate the use of probabilistic programming to superimpose protein structures guided by a Bayesian model. Our model THESEUS-PP is based on the THESEUS model, a probabilistic model of protein superposition based on rotation, translation and perturbation of an underlying, latent mean structure. The model was implemented in the probabilistic programming language Pyro. Unlike conventional methods that minimize the sum of the squared distances, THESEUS takes into account correlated atom positions and heteroscedasticity (ie. atom positions can feature different variances). THESEUS performs maximum likelihood estimation using iterative expectation-maximization. In contrast, THESEUS-PP allows automated maximum a-posteriori (MAP) estimation using suitable priors over rotation, translation, variances and latent mean structure. The results indicate that probabilistic programming is a powerful new paradigm for the formulation of Bayesian probabilistic models concerning biomolecular structure. Specifically, we envision the use of the THESEUS-PP model as a suitable error model or likelihood in Bayesian protein structure prediction using deep probabilistic programming.
Keywords: Bayesian modelling; deep probabilistic programming; protein structure prediction; protein superposition.
Figures


References
-
- Kabsch W, “A discussion of the solution for the best rotation to relate two sets of vectors,” Acta Cryst. A, vol. 34, pp. 827–828, 1978.
-
- Horn B, “Closed-form solution of absolute orientation using unit quaternions,” J. Opt. Soc. Am. A, vol. 4, pp. 629–642, 1987.
-
- Coutsias E, Seok C, and Dill K, “Using quaternions to calculate rmsd,” J. Comp. Chem, vol. 25, pp. 1849–1857, 2004. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
