Network potential identifies therapeutic miRNA cocktails in Ewing sarcoma
- PMID: 34662337
- PMCID: PMC8601628
- DOI: 10.1371/journal.pcbi.1008755
Network potential identifies therapeutic miRNA cocktails in Ewing sarcoma
Abstract
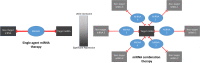
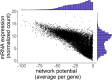
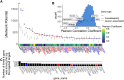
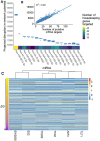
MicroRNA (miRNA)-based therapies are an emerging class of targeted therapeutics with many potential applications. Ewing Sarcoma patients could benefit dramatically from personalized miRNA therapy due to inter-patient heterogeneity and a lack of druggable (to this point) targets. However, because of the broad effects miRNAs may have on different cells and tissues, trials of miRNA therapies have struggled due to severe toxicity and unanticipated immune response. In order to overcome this hurdle, a network science-based approach is well-equipped to evaluate and identify miRNA candidates and combinations of candidates for the repression of key oncogenic targets while avoiding repression of essential housekeeping genes. We first characterized 6 Ewing sarcoma cell lines using mRNA sequencing. We then estimated a measure of tumor state, which we term network potential, based on both the mRNA gene expression and the underlying protein-protein interaction network in the tumor. Next, we ranked mRNA targets based on their contribution to network potential. We then identified miRNAs and combinations of miRNAs that preferentially act to repress mRNA targets with the greatest influence on network potential. Our analysis identified TRIM25, APP, ELAV1, RNF4, and HNRNPL as ideal mRNA targets for Ewing sarcoma therapy. Using predicted miRNA-mRNA target mappings, we identified miR-3613-3p, let-7a-3p, miR-300, miR-424-5p, and let-7b-3p as candidate optimal miRNAs for preferential repression of these targets. Ultimately, our work, as exemplified in the case of Ewing sarcoma, describes a novel pipeline by which personalized miRNA cocktails can be designed to maximally perturb gene networks contributing to cancer progression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

