High-risk subtypes of chronic lymphocytic leukemia are detectable as early as 16 years prior to diagnosis
- PMID: 34662377
- PMCID: PMC10650964
- DOI: 10.1182/blood.2021012890
High-risk subtypes of chronic lymphocytic leukemia are detectable as early as 16 years prior to diagnosis
Abstract
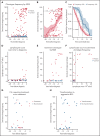
Chronic lymphocytic leukemia (CLL) is preceded by monoclonal B-cell lymphocytosis (MBL), a CLL precursor state with a prevalence of up to 12% in aged individuals; however, the duration of MBL and the mechanisms of its evolution to CLL remain largely unknown. In this study, we sequenced the B-cell receptor (BcR) immunoglobulin heavy chain (IGH) gene repertoire of 124 patients with CLL and 118 matched controls in blood samples taken up to 22 years prior to diagnosis. Significant skewing in the BcR IGH gene repertoire was detected in the majority of patients, even before the occurrence of lymphocytosis and irrespective of the clonotypic IGH variable gene somatic hypermutation status. Furthermore, we identified dominant clonotypes belonging to major stereotyped subsets associated with poor prognosis up to 16 years before diagnosis in 14 patients with CLL. In 22 patients with longitudinal samples, the skewing of the BcR IGH gene repertoire increased significantly over time to diagnosis or remained stable at high levels. For 14 of 16 patients with available samples at diagnosis, the CLL clonotype was already present in the prediagnostic samples. Overall, our data indicate that the preclinical phase of CLL could be longer than previously thought, even in adverse-prognostic cases.
© 2022 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Figures

Comment in
-
Premonoclonal B-cell lymphocytosis: the CLL cell of origin.Blood. 2022 Mar 10;139(10):1436-1437. doi: 10.1182/blood.2021014339. Blood. 2022. PMID: 35267006 No abstract available.
References
-
- Hallek M, Cheson BD, Catovsky D, et al. International Workshop on Chronic Lymphocytic Leukemia Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. - PMC - PubMed
-
- Hallek M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am J Hematol. 2019;94(11):1266–1287. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

