Positive Selection and Horizontal Gene Transfer in the Genome of a Male-Killing Wolbachia
- PMID: 34662426
- PMCID: PMC8763111
- DOI: 10.1093/molbev/msab303
Positive Selection and Horizontal Gene Transfer in the Genome of a Male-Killing Wolbachia
Abstract
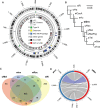
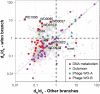
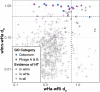
Wolbachia are a genus of widespread bacterial endosymbionts in which some strains can hijack or manipulate arthropod host reproduction. Male killing is one such manipulation in which these maternally transmitted bacteria benefit surviving daughters in part by removing competition with the sons for scarce resources. Despite previous findings of interesting genome features of microbial sex ratio distorters, the population genomics of male-killers remain largely uncharacterized. Here, we uncover several unique features of the genome and population genomics of four Arizonan populations of a male-killing Wolbachia strain, wInn, that infects mushroom-feeding Drosophila innubila. We first compared the wInn genome with other closely related Wolbachia genomes of Drosophila hosts in terms of genome content and confirm that the wInn genome is largely similar in overall gene content to the wMel strain infecting D. melanogaster. However, it also contains many unique genes and repetitive genetic elements that indicate lateral gene transfers between wInn and non-Drosophila eukaryotes. We also find that, in line with literature precedent, genes in the Wolbachia prophage and Octomom regions are under positive selection. Of all the genes under positive selection, many also show evidence of recent horizontal transfer among Wolbachia symbiont genomes. These dynamics of selection and horizontal gene transfer across the genomes of several Wolbachia strains and diverse host species may be important underlying factors in Wolbachia's success as a male-killer of divergent host species.
Keywords: Wolbachia; evolutionary genomics; male killing.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. - PubMed
-
- Baldo L, Ayoub NA, Hayashi CY, Russell JA, Stahlhut JK, Werren JH.. 2008. Insight into the routes of Wolbachia invasion: high levels of horizontal transfer in the spider genus Agelenopsis revealed by Wolbachia strain and mitochondrial DNA diversity. Mol Ecol. 17:557–569. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

