A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae
- PMID: 34663323
- PMCID: PMC8522093
- DOI: 10.1186/s12934-021-01691-3
A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae
Abstract
Background: Saccharomyces cerevisiae is an important synthetic biology chassis for microbial production of valuable molecules. Promoter engineering has been frequently applied to generate more synthetic promoters with a variety of defined characteristics in order to achieve a well-regulated genetic network for high production efficiency. Galactose-inducible (GAL) expression systems, composed of GAL promoters and multiple GAL regulators, have been widely used for protein overexpression and pathway construction in S. cerevisiae. However, the function of each element in synthetic promoters and how they interact with GAL regulators are not well known.
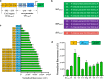
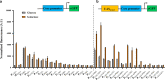
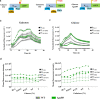
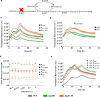
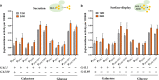
Results: Here, a library of synthetic GAL promoters demonstrate that upstream activating sequences (UASs) and core promoters have a synergistic relationship that determines the performance of each promoter under different carbon sources. We found that the strengths of synthetic GAL promoters could be fine-tuned by manipulating the sequence, number, and substitution of UASs. Core promoter replacement generated synthetic promoters with a twofold strength improvement compared with the GAL1 promoter under multiple different carbon sources in a strain with GAL1 and GAL80 engineering. These results represent an expansion of the classic GAL expression system with an increased dynamic range and a good tolerance of different carbon sources.
Conclusions: In this study, the effect of each element on synthetic GAL promoters has been evaluated and a series of well-controlled synthetic promoters are constructed. By studying the interaction of synthetic promoters and GAL regulators, synthetic promoters with an increased dynamic range under different carbon sources are created.
Keywords: Carbon sources; Protein expression; Saccharomyces cerevisiae; Synthetic promoter.
© 2021. The Author(s).
Conflict of interest statement
X.L. has a financial interest in Demetrix. J.D.K. has a financial interest in Amyris, Lygos, Demetrix, Napigen, Maple Bio, Apertor Labs, Zero Acre Farms, Berkeley Yeast, and Ansa Biotechnology.
Figures
Similar articles
-
Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuac029. doi: 10.1093/jimb/kuac029. J Ind Microbiol Biotechnol. 2023. PMID: 36633543 Free PMC article. Review.
-
Promoters inducible by aromatic amino acids and γ-aminobutyrate (GABA) for metabolic engineering applications in Saccharomyces cerevisiae.Appl Microbiol Biotechnol. 2015 Mar;99(6):2705-14. doi: 10.1007/s00253-014-6303-5. Epub 2015 Jan 10. Appl Microbiol Biotechnol. 2015. PMID: 25573467
-
An Expanded Heterologous GAL Promoter Collection for Diauxie-Inducible Expression in Saccharomyces cerevisiae.ACS Synth Biol. 2018 Feb 16;7(2):748-751. doi: 10.1021/acssynbio.7b00355. Epub 2018 Jan 23. ACS Synth Biol. 2018. PMID: 29301066
-
Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters.Biotechnol Bioeng. 2012 Nov;109(11):2884-95. doi: 10.1002/bit.24552. Epub 2012 May 17. Biotechnol Bioeng. 2012. PMID: 22565375
-
From natural to synthetic: Promoter engineering in yeast expression systems.Biotechnol Adv. 2024 Dec;77:108446. doi: 10.1016/j.biotechadv.2024.108446. Epub 2024 Sep 7. Biotechnol Adv. 2024. PMID: 39245291 Review.
Cited by
-
Engineering artificial cross-species promoters with different transcriptional strengths.Synth Syst Biotechnol. 2024 Aug 8;10(1):49-57. doi: 10.1016/j.synbio.2024.08.003. eCollection 2025. Synth Syst Biotechnol. 2024. PMID: 39224149 Free PMC article.
-
The kinase Rio1 and a ribosome collision-dependent decay pathway survey the integrity of 18S rRNA cleavage.PLoS Biol. 2024 Apr 25;22(4):e3001767. doi: 10.1371/journal.pbio.3001767. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 39038273 Free PMC article.
-
Design of synthetic promoters for cyanobacteria with generative deep-learning model.Nucleic Acids Res. 2023 Jul 21;51(13):7071-7082. doi: 10.1093/nar/gkad451. Nucleic Acids Res. 2023. PMID: 37246641 Free PMC article.
-
Cyanamide-inducible expression of homing nuclease I- SceI for selectable marker removal and promoter characterisation in Saccharomyces cerevisiae.Synth Syst Biotechnol. 2024 Jun 28;9(4):820-827. doi: 10.1016/j.synbio.2024.06.009. eCollection 2024 Dec. Synth Syst Biotechnol. 2024. PMID: 39072146 Free PMC article.
-
Natural promoters and promoter engineering strategies for metabolic regulation in Saccharomyces cerevisiae.J Ind Microbiol Biotechnol. 2023 Feb 17;50(1):kuac029. doi: 10.1093/jimb/kuac029. J Ind Microbiol Biotechnol. 2023. PMID: 36633543 Free PMC article. Review.
References
-
- Turanlı-Yıldız B, Hacısalihoğlu B, Çakar ZP. Advances in metabolic engineering of Saccharomyces cerevisiae for the production of industrially and clinically important chemicals. Old Yeasts New Quest. 2017 doi: 10.5772/intechopen.70327. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

