Parainfluenza virus entry at the onset of infection
- PMID: 34663496
- PMCID: PMC10270304
- DOI: 10.1016/bs.aivir.2021.07.001
Parainfluenza virus entry at the onset of infection
Abstract
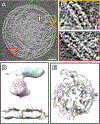
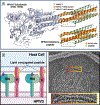
Parainfluenza viruses, members of the enveloped, negative-sense, single stranded RNA Paramyxoviridae family, impact global child health as the cause of significant lower respiratory tract infections. Parainfluenza viruses enter cells by fusing directly at the cell surface membrane. How this fusion occurs via the coordinated efforts of the two molecules that comprise the viral surface fusion complex, and how these efforts may be blocked, are the subjects of this chapter. The receptor binding protein of parainfluenza forms a complex with the fusion protein of the virus, remaining stably associated until a receptor is reached. At that point, the receptor binding protein actively triggers the fusion protein to undergo a series of transitions that ultimately lead to membrane fusion and viral entry. In recent years it has become possible to examine this remarkable process on the surface of viral particles and to begin to understand the steps in the transition of this molecular machine, using a structural biology approach. Understanding the steps in entry leads to several possible strategies to prevent fusion and inhibit infection.
Keywords: Antivirals; Cryo-electron tomography; Fusion inhibitors; Membrane fusion; Parainfluenza virus; Viral entry.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures



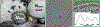


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

