Noise2Void: unsupervised denoising of PET images
- PMID: 34663767
- PMCID: PMC8563445
- DOI: 10.1088/1361-6560/ac30a0
Noise2Void: unsupervised denoising of PET images
Abstract
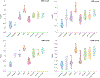
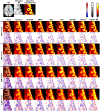
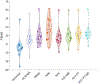
Objective:Elevated noise levels in positron emission tomography (PET) images lower image quality and quantitative accuracy and are a confounding factor for clinical interpretation. The objective of this paper is to develop a PET image denoising technique based on unsupervised deep learning.Significance:Recent advances in deep learning have ushered in a wide array of novel denoising techniques, several of which have been successfully adapted for PET image reconstruction and post-processing. The bulk of the deep learning research so far has focused on supervised learning schemes, which, for the image denoising problem, require paired noisy and noiseless/low-noise images. This requirement tends to limit the utility of these methods for medical applications as paired training datasets are not always available. Furthermore, to achieve the best-case performance of these methods, it is essential that the datasets for training and subsequent real-world application have consistent image characteristics (e.g. noise, resolution, etc), which is rarely the case for clinical data. To circumvent these challenges, it is critical to develop unsupervised techniques that obviate the need for paired training data.Approach:In this paper, we have adapted Noise2Void, a technique that relies on corrupt images alone for model training, for PET image denoising and assessed its performance using PET neuroimaging data. Noise2Void is an unsupervised approach that uses a blind-spot network design. It requires only a single noisy image as its input, and, therefore, is well-suited for clinical settings. During the training phase, a single noisy PET image serves as both the input and the target. Here we present a modified version of Noise2Void based on a transfer learning paradigm that involves group-level pretraining followed by individual fine-tuning. Furthermore, we investigate the impact of incorporating an anatomical image as a second input to the network.Main Results:We validated our denoising technique using simulation data based on the BrainWeb digital phantom. We show that Noise2Void with pretraining and/or anatomical guidance leads to higher peak signal-to-noise ratios than traditional denoising schemes such as Gaussian filtering, anatomically guided non-local means filtering, and block-matching and 4D filtering. We used the Noise2Noise denoising technique as an additional benchmark. For clinical validation, we applied this method to human brain imaging datasets. The clinical findings were consistent with the simulation results confirming the translational value of Noise2Void as a denoising tool.
Keywords: PET; deep learning; denoising; unsupervised learning.
© 2021 Institute of Physics and Engineering in Medicine.
Figures

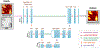





References
-
- Arabi H and Zaidi H 2020. Spatially guided nonlocal mean approach for denoising of PET images Med Phys 47(4), 1656–1669. - PubMed
-
- Bergmann S, Fox K, Rand A, McElvany K, Welch M, Markham J and Sobel B 1984. Quantification of regional myocardial blood flow in vivo with Circulation 70(4), 724–733. - PubMed
-
- Boussion N, Cheze Le Rest C, Hatt M and Visvikis D 2009. Incorporation of wavelet-based denoising in iterative deconvolution for partial volume correction in whole-body PET imaging Eur J Nucl Med Mol Imaging 36(7), 1064–1075. - PubMed
-
- Buades A, Coll B and Morel JM 2005. A non-local algorithm for image denoising in ‘Computer Vision and Pattern Recognition, IEEE Computer Society Conference on’ Vol. 2 pp. 60–65.
-
- Chan C, Fulton R, Barnett R, Feng DD and Meikle S 2014. Postreconstruction nonlocal means filtering of whole-body PET with an anatomical prior IEEE Trans Med Imaging 33(3), 636–650. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
