Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy
- PMID: 34663807
- PMCID: PMC8524723
- DOI: 10.1038/s41467-021-26282-z
Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy
Abstract
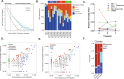
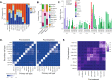
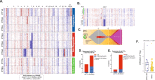
In contrast to the curative effect of allogenic stem cell transplantation in acute myeloid leukemia via T cell activity, only modest responses are achieved with checkpoint-blockade therapy, which might be explained by T cell phenotypes and T cell receptor (TCR) repertoires. Here, we show by paired single-cell RNA analysis and TCR repertoire profiling of bone marrow cells in relapsed/refractory acute myeloid leukemia patients pre/post azacytidine+nivolumab treatment that the disease-related T cell subsets are highly heterogeneous, and their abundance changes following PD-1 blockade-based treatment. TCR repertoires expand and primarily emerge from CD8+ cells in patients responding to treatment or having a stable disease, while TCR repertoires contract in therapy-resistant patients. Trajectory analysis reveals a continuum of CD8+ T cell phenotypes, characterized by differential expression of granzyme B and a bone marrow-residing memory CD8+ T cell subset, in which a population with stem-like properties expressing granzyme K is enriched in responders. Chromosome 7/7q loss, on the other hand, is a cancer-intrinsic genomic marker of PD-1 blockade resistance in AML. In summary, our study reveals that adaptive T cell plasticity and genomic alterations determine responses to PD-1 blockade in acute myeloid leukemia.
© 2021. The Author(s).
Conflict of interest statement
K.Ta. reports consulting and advisory roles for Symbio Pharmaceuticals, Novartis, GSK and Celgene/BMS. G.A. reports consulting fees from Novartis and Poseida therapeutics, and research funding from Merck and Jenssen Pharmaceuticals. M.R.G. reports consulting fees with VeraStem Oncology and stock/ownership interest KDAc Therapeutics. M.K. reports grant support and consulting fees from AbbVie, Genentech, F. Hoffmann La-Roche, Stemline Therapeutics, Forty-Seven, consulting fees from Amgen and Kisoji, grant support from Eli Lilly, Cellectis, Calithera, Ablynx, Agios, Ascentage, AstraZeneca, Rafael Pharmaceutical, Sanofi, royalties and stock options from Reata Pharmaceutical Inc. P.S. reports consulting, advisory roles, and/or stocks/ownership for Achelois, Adaptive Biotechnologies, Affini-T, Apricity, BioAtla, BioNTech, Candel Therapeutics, Catalio, Codiak, Constellation, Dragonfly, Earli, Enable Medicine, Glympse, Hummingbird, ImaginAb, Infinity Pharma, Jounce, JSL Health, Lava Therapeutics, Lytix, Marker, Oncolytics, PBM Capital, Phenomic AI, Polaris Pharma, Sporos, Time Bioventures, Trained Therapeutix, Two Bear Capital, Venn Biosciences. J.P.A. reports consulting, advisory roles, and/or stocks/ownership for Achelois, Adaptive Biotechnologies, Apricity, BioAtla, BioNTech, Candel Therapeutics, Codiak, Dragonfly, Earli, Enable Medicine, Hummingbird, ImaginAb, Jounce, Lava Therapeutics, Lytix, Marker, PBM Capital, Phenomic AI, Polaris Pharma, Time Bioventures, Trained Therapeutix, Two Bear Capital, Venn Biosciences. N.D. reports research funding from Daiichi Sankyo, Bristol-Myers Squibb, Pfizer, Karyopharm, Sevier, Genentech, Astellas, Abbvie, Genentech, Novimmune, Amgen, Trovagene, Gilead and ImmunoGen and has served in a consulting or advisory role for Daiichi Sankyo, Bristol-Myers Squibb, Pfizer, Novartis, Celgene, AbbVie, Genentech, Servier, Trillium, Syndax, Trovagene, Astellas, Gilead and Agios. The remaining authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

