Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma
- PMID: 34663816
- PMCID: PMC8523550
- DOI: 10.1038/s41467-021-26343-3
Single-cell transcriptomic analysis of the tumor ecosystems underlying initiation and progression of papillary thyroid carcinoma
Abstract
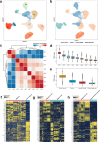
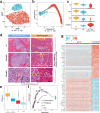
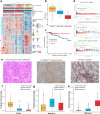
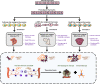
The tumor ecosystem of papillary thyroid carcinoma (PTC) is poorly characterized. Using single-cell RNA sequencing, we profile transcriptomes of 158,577 cells from 11 patients' paratumors, localized/advanced tumors, initially-treated/recurrent lymph nodes and radioactive iodine (RAI)-refractory distant metastases, covering comprehensive clinical courses of PTC. Our data identifies a "cancer-primed" premalignant thyrocyte population with normal morphology but altered transcriptomes. Along the developmental trajectory, we also discover three phenotypes of malignant thyrocytes (follicular-like, partial-epithelial-mesenchymal-transition-like, dedifferentiation-like), whose composition shapes bulk molecular subtypes, tumor characteristics and RAI responses. Furthermore, we uncover a distinct BRAF-like-B subtype with predominant dedifferentiation-like thyrocytes, enriched cancer-associated fibroblasts, worse prognosis and promising prospect of immunotherapy. Moreover, potential vascular-immune crosstalk in PTC provides theoretical basis for combined anti-angiogenic and immunotherapy. Together, our findings provide insight into the PTC ecosystem that suggests potential prognostic and therapeutic implications.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

