Dissecting single-cell genomes through the clonal organoid technique
- PMID: 34663940
- PMCID: PMC8569207
- DOI: 10.1038/s12276-021-00680-1
Dissecting single-cell genomes through the clonal organoid technique
Abstract
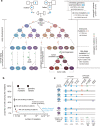
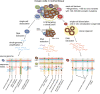
The revolution in genome sequencing technologies has enabled the comprehensive detection of genomic variations in human cells, including inherited germline polymorphisms, de novo mutations, and postzygotic mutations. When these technologies are combined with techniques for isolating and expanding single-cell DNA, the landscape of somatic mosaicism in an individual body can be systematically revealed at a single-cell resolution. Here, we summarize three strategies (whole-genome amplification, microdissection of clonal patches in the tissue, and in vitro clonal expansion of single cells) that are currently applied for single-cell mutational analyses. Among these approaches, in vitro clonal expansion, particularly via adult stem cell-derived organoid culture technologies, yields the most sensitive and precise catalog of somatic mutations in single cells. Moreover, because it produces living mutant cells, downstream validation experiments and multiomics profiling are possible. Through the synergistic combination of organoid culture and genome sequencing, researchers can track genome changes at a single-cell resolution, which will lead to new discoveries that were previously impossible.
© 2021. The Author(s).
Conflict of interest statement
Young Seok Ju is the founder and CEO of GENOME INSIGHT Inc.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

