Harnessing multimodal data integration to advance precision oncology
- PMID: 34663944
- PMCID: PMC8810682
- DOI: 10.1038/s41568-021-00408-3
Harnessing multimodal data integration to advance precision oncology
Abstract
Advances in quantitative biomarker development have accelerated new forms of data-driven insights for patients with cancer. However, most approaches are limited to a single mode of data, leaving integrated approaches across modalities relatively underdeveloped. Multimodal integration of advanced molecular diagnostics, radiological and histological imaging, and codified clinical data presents opportunities to advance precision oncology beyond genomics and standard molecular techniques. However, most medical datasets are still too sparse to be useful for the training of modern machine learning techniques, and significant challenges remain before this is remedied. Combined efforts of data engineering, computational methods for analysis of heterogeneous data and instantiation of synergistic data models in biomedical research are required for success. In this Perspective, we offer our opinions on synthesizing complementary modalities of data with emerging multimodal artificial intelligence methods. Advancing along this direction will result in a reimagined class of multimodal biomarkers to propel the field of precision oncology in the coming decade.
© 2021. Springer Nature Limited.
Figures
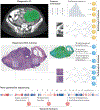

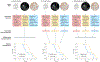

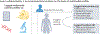

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

