Retrospective analysis of Schlafen11 (SLFN11) to predict the outcomes to therapies affecting the DNA damage response
- PMID: 34663950
- PMCID: PMC8651811
- DOI: 10.1038/s41416-021-01560-1
Retrospective analysis of Schlafen11 (SLFN11) to predict the outcomes to therapies affecting the DNA damage response
Abstract
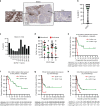
Background: The absence of the putative DNA/RNA helicase Schlafen11 (SLFN11) is thought to cause resistance to DNA-damaging agents (DDAs) and PARP inhibitors.
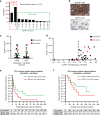
Methods: We developed and validated a clinically applicable SLFN11 immunohistochemistry assay and retrospectively correlated SLFN11 tumour levels to patient outcome to the standard of care therapies and olaparib maintenance.
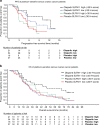
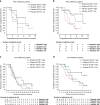
Results: High SLFN11 associated with improved prognosis to the first-line treatment with DDAs platinum-plus-etoposide in SCLC patients, but was not strongly linked to paclitaxel-platinum response in ovarian cancer patients. Multivariate analysis of patients with relapsed platinum-sensitive ovarian cancer from the randomised, placebo-controlled Phase II olaparib maintenance Study19 showed SLFN11 tumour levels associated with sensitivity to olaparib. Study19 patients with high SLFN11 had a lower progression-free survival (PFS) hazard ratio compared to patients with low SLFN11, although both groups had the benefit of olaparib over placebo. Whilst caveated by small sample size, this trend was maintained for PFS, but not overall survival, when adjusting for BRCA status across the olaparib and placebo treatment groups, a key driver of PARP inhibitor sensitivity.
Conclusion: We provide clinical evidence supporting the role of SLFN11 as a DDA therapy selection biomarker in SCLC and highlight the need for further clinical investigation into SLFN11 as a PARP inhibitor predictive biomarker.
© 2021. The Author(s).
Conflict of interest statement
SEW, CW, MPR, TB, PMC, EVJ, PR, JRC, HKA, FSLN, PMW, DH, ED, EAH, JCB, AJP, EL & GNJ were employees of AstraZeneca during this study. All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

