Circadian clocks guide dendritic cells into skin lymphatics
- PMID: 34663979
- PMCID: PMC8553624
- DOI: 10.1038/s41590-021-01040-x
Circadian clocks guide dendritic cells into skin lymphatics
Abstract
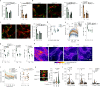
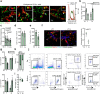
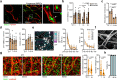
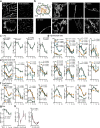
Migration of leukocytes from the skin to lymph nodes (LNs) via afferent lymphatic vessels (LVs) is pivotal for adaptive immune responses1,2. Circadian rhythms have emerged as important regulators of leukocyte trafficking to LNs via the blood3,4. Here, we demonstrate that dendritic cells (DCs) have a circadian migration pattern into LVs, which peaks during the rest phase in mice. This migration pattern is determined by rhythmic gradients in the expression of the chemokine CCL21 and of adhesion molecules in both mice and humans. Chronopharmacological targeting of the involved factors abrogates circadian migration of DCs. We identify cell-intrinsic circadian oscillations in skin lymphatic endothelial cells (LECs) and DCs that cogovern these rhythms, as their genetic disruption in either cell type ablates circadian trafficking. These observations indicate that circadian clocks control the infiltration of DCs into skin lymphatics, a process that is essential for many adaptive immune responses and relevant for vaccination and immunotherapies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

