Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe-host-communication in vivo
- PMID: 34664784
- PMCID: PMC8524437
- DOI: 10.1002/jev2.12159
Visualizing transfer of microbial biomolecules by outer membrane vesicles in microbe-host-communication in vivo
Abstract
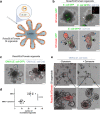
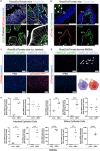
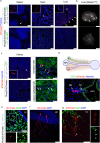
The intestinal microbiota influences mammalian host physiology in health and disease locally in the gut but also in organs devoid of direct contact with bacteria such as the liver and brain. Extracellular vesicles (EVs) or outer membrane vesicles (OMVs) released by microbes are increasingly recognized for their potential role as biological shuttle systems for inter-kingdom communication. However, physiologically relevant evidence for the transfer of functional biomolecules from the intestinal microbiota to individual host cells by OMVs in vivo is scarce. By introducing Escherichia coli engineered to express Cre-recombinase (E. coliCre ) into mice with a Rosa26.tdTomato-reporter background, we leveraged the Cre-LoxP system to report the transfer of bacterial OMVs to recipient cells in vivo. Colonizing the intestine of these mice with E. coliCre , resulted in Cre-recombinase induced fluorescent reporter gene-expression in cells along the intestinal epithelium, including intestinal stem cells as well as mucosal immune cells such as macrophages. Furthermore, even far beyond the gut, bacterial-derived Cre induced extended marker gene expression in a wide range of host tissues, including the heart, liver, kidney, spleen, and brain. Together, our findings provide a method and proof of principle that OMVs can serve as a biological shuttle system for the horizontal transfer of functional biomolecules between bacteria and mammalian host cells.
Keywords: Cre-Loxp; extracellular vesicles; inflammation; intestinal stem cells; microbiome; outer membrane vesicles.
© 2021 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals, LLC on behalf of the International Society for Extracellular Vesicles.
Conflict of interest statement
Authors declare no conflict of interest.
Figures


References
-
- Albillos, A. , de Gottardi, A. , & Rescigno, M. (2020). The gut‐liver axis in liver disease: Pathophysiological basis for therapy. Journal of Hepatology 72, 558–577. - PubMed
-
- Barbier, M. , & Mittar, D. (2014). Novel fluorescent reporters for studying host‐pathogen interactions. ATCC Application Note.1–6.
-
- Barker, N. , van Es, J. H. , Kuipers, J. , Kujala, P. , van den Born, M. , Cozijnsen, M. , Haegebarth, A. , Korving, J. , Begthel, H. , Peters, P. J. , & Clevers, H. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007. - PubMed
-
- Becker, C. , Wirtz, S. , Blessing, M. , Pirhonen, J. , Strand, D. , Bechthold, O. , Frick, J. , Galle, P. R. , Autenrieth, I. , & Neurath, M. F. (2003). Constitutive p40 promoter activation and IL‐23 production in the terminal ileum mediated by dendritic cells. Journal of Clinical Investigation 112, 693–706. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

