Human CEACAM1-LF regulates lipid storage in HepG2 cells via fatty acid transporter CD36
- PMID: 34666041
- PMCID: PMC8577156
- DOI: 10.1016/j.jbc.2021.101311
Human CEACAM1-LF regulates lipid storage in HepG2 cells via fatty acid transporter CD36
Abstract
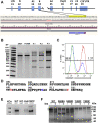
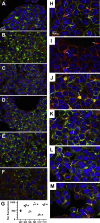
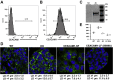
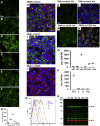
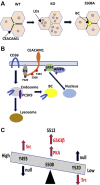
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) is expressed in the liver and secreted as biliary glycoprotein 1 (BGP1) via bile canaliculi (BCs). CEACAM1-LF is a 72 amino acid cytoplasmic domain mRNA splice isoform with two immunoreceptor tyrosine-based inhibitory motifs (ITIMs). Ceacam1-/- or Ser503Ala transgenic mice have been shown to develop insulin resistance and nonalcoholic fatty liver disease; however, the role of the human equivalent residue, Ser508, in lipid dysregulation is unknown. Human HepG2 hepatocytes that express CEACAM1 and form BC in vitro were compared with CEACAM1-/- cells and CEACAM1-/- cells expressing Ser508Ala null or Ser508Asp phosphorylation mimic mutations or to phosphorylation null mutations in the tyrosine ITIMs known to be phosphorylated by the tyrosine kinase Src. CEACAM1-/- cells and the Ser508Asp and Tyr520Phe mutants strongly retained lipids, while Ser508Ala and Tyr493Phe mutants had low lipid levels compared with wild-type cells, indicating that the ITIM mutants phenocopied the Ser508 mutants. We found that the fatty acid transporter CD36 was upregulated in the S508A mutant, coexpressed in BCs with CEACAM1, co-IPed with CEACAM1 and Src, and when downregulated via RNAi, an increase in lipid droplet content was observed. Nuclear translocation of CD36 associated kinase LKB1 was increased sevenfold in the S508A mutant versus CEACAM1-/- cells and correlated with increased activation of CD36-associated kinase AMPK in CEACAM1-/- cells. Thus, while CEACAM1-/- HepG2 cells upregulate lipid storage similar to Ceacam1-/- in murine liver, the null mutation Ser508Ala led to decreased lipid storage, emphasizing evolutionary changes between the CEACAM1 genes in mouse and humans.
Keywords: CD36; CEACAM1; NAFLD; NASH; bile canaliculi; lipid storage.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Beauchemin N., Draber P., Dveksler G., Gold P., Gray-Owen S., Grunert F., Hammarstrom S., Holmes K.V., Karlsson A., Kuroki M., Lin S.H., Lucka L., Najjar S.M., Neumaier M., Obrink B. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 1999;252:243–249. - PubMed
-
- Poy M.N., Yang Y., Rezaei K., Fernstrom M.A., Lee A.D., Kido Y., Erickson S.K., Najjar S.M. CEACAM1 regulates insulin clearance in liver. Nat. Genet. 2002;30:270–276. - PubMed
-
- Jirsa M., Muchova L., Draberova L., Draber P., Smid F., Kuroki M., Marecek Z., Groen A.K. Carcinoembryonic antigen-related cell adhesion molecule 1 is the 85-kilodalton pronase-resistant biliary glycoprotein in the cholesterol crystallization promoting low density protein-lipid complex. Hepatology. 2001;34:1075–1082. - PubMed
-
- Kitamura Y., Murata Y., Park J.H., Kotani T., Imada S., Saito Y., Okazawa H., Azuma T., Matozaki T. Regulation by gut commensal bacteria of carcinoembryonic antigen-related cell adhesion molecule expression in the intestinal epithelium. Genes Cells. 2015;20:578–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

