Transcription and splicing dynamics during early Drosophila development
- PMID: 34667107
- PMCID: PMC8906543
- DOI: 10.1261/rna.078933.121
Transcription and splicing dynamics during early Drosophila development
Abstract
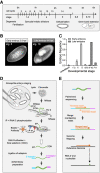
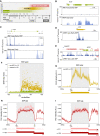
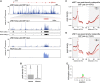
Widespread cotranscriptional splicing has been demonstrated from yeast to human. However, most studies to date addressing the kinetics of splicing relative to transcription used either Saccharomyces cerevisiae or metazoan cultured cell lines. Here, we adapted native elongating transcript sequencing technology (NET-seq) to measure cotranscriptional splicing dynamics during the early developmental stages of Drosophila melanogaster embryos. Our results reveal the position of RNA polymerase II (Pol II) when both canonical and recursive splicing occur. We found heterogeneity in splicing dynamics, with some RNAs spliced immediately after intron transcription, whereas for other transcripts no splicing was observed over the first 100 nt of the downstream exon. Introns that show splicing completion before Pol II has reached the end of the downstream exon are necessarily intron-defined. We studied the splicing dynamics of both nascent pre-mRNAs transcribed in the early embryo, which have few and short introns, as well as pre-mRNAs transcribed later in embryonic development, which contain multiple long introns. As expected, we found a relationship between the proportion of spliced reads and intron size. However, intron definition was observed at all intron sizes. We further observed that genes transcribed in the early embryo tend to be isolated in the genome whereas genes transcribed later are often overlapped by a neighboring convergent gene. In isolated genes, transcription termination occurred soon after the polyadenylation site, while in overlapped genes, Pol II persisted associated with the DNA template after cleavage and polyadenylation of the nascent transcript. Taken together, our data unravel novel dynamic features of Pol II transcription and splicing in the developing Drosophila embryo.
Keywords: Drosophila melanogaster embryo; NET-seq; splicing kinetics; transcription termination.
© 2022 Prudêncio et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures







Similar articles
-
Splicing Kinetics and Coordination Revealed by Direct Nascent RNA Sequencing through Nanopores.Mol Cell. 2020 Mar 5;77(5):985-998.e8. doi: 10.1016/j.molcel.2019.11.017. Epub 2019 Dec 12. Mol Cell. 2020. PMID: 31839405 Free PMC article.
-
Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II.Cell. 2016 Apr 7;165(2):372-381. doi: 10.1016/j.cell.2016.02.045. Epub 2016 Mar 24. Cell. 2016. PMID: 27020755 Free PMC article.
-
Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse.RNA. 2012 Dec;18(12):2174-86. doi: 10.1261/rna.034090.112. Epub 2012 Oct 24. RNA. 2012. PMID: 23097425 Free PMC article.
-
Pre-mRNA splicing and its cotranscriptional connections.Trends Genet. 2023 Sep;39(9):672-685. doi: 10.1016/j.tig.2023.04.008. Epub 2023 May 24. Trends Genet. 2023. PMID: 37236814 Free PMC article. Review.
-
Extremely fast and incredibly close: cotranscriptional splicing in budding yeast.RNA. 2017 May;23(5):601-610. doi: 10.1261/rna.060830.117. Epub 2017 Feb 2. RNA. 2017. PMID: 28153948 Free PMC article. Review.
Cited by
-
Optogenetic manipulation of nuclear Dorsal reveals temporal requirements and consequences for transcription.Development. 2025 Mar 15;152(6):dev204706. doi: 10.1242/dev.204706. Epub 2025 Mar 31. Development. 2025. PMID: 40018801 Free PMC article.
-
Revealing the hidden RBP-RNA interactions with RNA modification enzyme-based strategies.Wiley Interdiscip Rev RNA. 2024 May-Jun;15(3):e1863. doi: 10.1002/wrna.1863. Wiley Interdiscip Rev RNA. 2024. PMID: 39392204 Free PMC article. Review.
-
RNA polymerase II is recruited to DNA double-strand breaks for dilncRNA transcription in Drosophila.RNA Biol. 2022;19(1):68-77. doi: 10.1080/15476286.2021.2014694. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 34965182 Free PMC article.
-
Combined modelling of mRNA decay dynamics and single-molecule imaging in the Drosophila embryo uncovers a role for P-bodies in 5' to 3' degradation.PLoS Biol. 2023 Jan 17;21(1):e3001956. doi: 10.1371/journal.pbio.3001956. eCollection 2023 Jan. PLoS Biol. 2023. PMID: 36649329 Free PMC article.
-
Transcriptomics in the era of long-read sequencing.Nat Rev Genet. 2025 Mar 28. doi: 10.1038/s41576-025-00828-z. Online ahead of print. Nat Rev Genet. 2025. PMID: 40155769 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
