XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer
- PMID: 34667145
- PMCID: PMC8526672
- DOI: 10.1038/s41392-021-00761-7
XBP1 regulates the protumoral function of tumor-associated macrophages in human colorectal cancer
Abstract
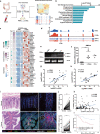
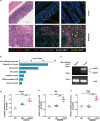
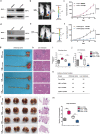
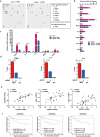
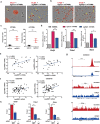
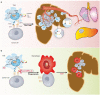
Macrophages are among the most abundant immune cells in colorectal cancer (CRC). Re-educating tumor-associated macrophages (TAMs) to switch from protumoral to anti-tumoral activity is an attractive treatment strategy that warrants further investigation. However, little is known about the key pathway that is activated in TAMs. In this study, infitrating CD206+ TAMs in CRC were sorted and subjected to RNA-seq analysis. Differentially expressed genes were found to be enriched in unfolded protein response/endoplasmic reticulum stress response processes, and XBP1 splicing/activation was specifically observed in TAMs. XBP1 activation in TAMs promoted the growth and metastasis of CRC. Ablation of XBP1 inhibited the expression of the pro-tumor cytokine signature of TAMs, including IL-6, VEGFA, and IL-4. Simultaneously, XBP1 depletion could directly inhibit the expression of SIRPα and THBS1, thereby blocking "don't eat me" recognition signals and enhancing phagocytosis. Therapeutic XBP1 gene editing using AAV2-sgXBP1 enhanced the anti-tumor activity. Together, XBP1 activation in TAMs drives CRC progression by elevating pro-tumor cytokine expression and secretion, as well as inhibiting macrophage phagocytosis. Targeting XBP1 signaling in TAMs may be a potential strategy for CRC therapy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Ap-2α/Elk-1 axis regulates Sirpα-dependent tumor phagocytosis by tumor-associated macrophages in colorectal cancer.Signal Transduct Target Ther. 2020 Apr 15;5(1):35. doi: 10.1038/s41392-020-0124-z. Signal Transduct Target Ther. 2020. PMID: 32296015 Free PMC article.
-
A STING agonist prodrug reprograms tumor-associated macrophage to boost colorectal cancer immunotherapy.Theranostics. 2025 Jan 1;15(1):277-299. doi: 10.7150/thno.101001. eCollection 2025. Theranostics. 2025. PMID: 39744236 Free PMC article.
-
X-box Binding Protein 1 Regulates Unfolded Protein, Acute-Phase, and DNA Damage Responses During Regeneration of Mouse Liver.Gastroenterology. 2017 Apr;152(5):1203-1216.e15. doi: 10.1053/j.gastro.2016.12.040. Epub 2017 Jan 9. Gastroenterology. 2017. PMID: 28082079
-
Natural compounds modulate the mechanism of action of tumour-associated macrophages against colorectal cancer: a review.J Cancer Res Clin Oncol. 2024 Nov 15;150(11):502. doi: 10.1007/s00432-024-06022-8. J Cancer Res Clin Oncol. 2024. PMID: 39546016 Free PMC article. Review.
-
Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis.Int J Mol Sci. 2021 Aug 6;22(16):8470. doi: 10.3390/ijms22168470. Int J Mol Sci. 2021. PMID: 34445193 Free PMC article. Review.
Cited by
-
Decoding endoplasmic reticulum stress signals in cancer cells and antitumor immunity.Trends Cancer. 2022 Nov;8(11):930-943. doi: 10.1016/j.trecan.2022.06.006. Epub 2022 Jul 8. Trends Cancer. 2022. PMID: 35817701 Free PMC article. Review.
-
The role of adipogenic niche resident cells in colorectal cancer progression in relation to obesity.Obes Rev. 2025 Apr;26(4):e13873. doi: 10.1111/obr.13873. Epub 2025 Jan 6. Obes Rev. 2025. PMID: 39763022 Free PMC article. Review.
-
Immunosuppressive Signaling Pathways as Targeted Cancer Therapies.Biomedicines. 2022 Mar 16;10(3):682. doi: 10.3390/biomedicines10030682. Biomedicines. 2022. PMID: 35327484 Free PMC article. Review.
-
The role and therapeutic implication of endoplasmic reticulum stress in inflammatory cancer transformation.Am J Cancer Res. 2022 May 15;12(5):2277-2292. eCollection 2022. Am J Cancer Res. 2022. PMID: 35693091 Free PMC article. Review.
-
How the Unfolded Protein Response Is a Boon for Tumors and a Bane for the Immune System.Immunohorizons. 2023 Apr 1;7(4):256-264. doi: 10.4049/immunohorizons.2200064. Immunohorizons. 2023. PMID: 37067519 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

