A dual-reporter system for investigating and optimizing protein translation and folding in E. coli
- PMID: 34667164
- PMCID: PMC8526717
- DOI: 10.1038/s41467-021-26337-1
A dual-reporter system for investigating and optimizing protein translation and folding in E. coli
Abstract
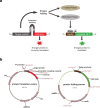
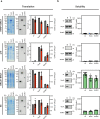
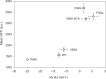
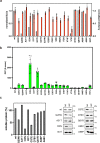
Strategies for investigating and optimizing the expression and folding of proteins for biotechnological and pharmaceutical purposes are in high demand. Here, we describe a dual-reporter biosensor system that simultaneously assesses in vivo protein translation and protein folding, thereby enabling rapid screening of mutant libraries. We have validated the dual-reporter system on five different proteins and find an excellent correlation between reporter signals and the levels of protein expression and solubility of the proteins. We further demonstrate the applicability of the dual-reporter system as a screening assay for deep mutational scanning experiments. The system enables high throughput selection of protein variants with high expression levels and altered protein stability. Next generation sequencing analysis of the resulting libraries of protein variants show a good correlation between computationally predicted and experimentally determined protein stabilities. We furthermore show that the mutational experimental data obtained using this system may be useful for protein structure calculations.
© 2021. The Author(s).
Conflict of interest statement
A.T.N., A.Z. and R.L. have filed a provisional application on this work (EP3209795B1, US10544414B2). The application covers the use of the two-cassette reporter system for assessing gene target translation and folding. All other authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

